- 1Division of Plant Science and Technology, University of Missouri, Columbia, MO, United States
- 2Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, MO, United States
- 3MU Data Science and Informatics Institute, University of Missouri, Columbia, MO, United States
- 4Department of Agronomy and Horticulture, University of Nebraska, Lincoln, NE, United States
- 5Department of Crop Sciences, University of Illinois, Urbana, IL, United States
- 6Department of Agronomy and Plant Genetics, University of Minnesota, Saint Paul, MN, United States
- 7Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, United States
- 8Department of Electrical Engineering and Computer Science, University of Missouri, Columbia, MO, United States
- 9Department of Health Management and Informatics, School of Medicine, University of Missouri, Columbia, MO, United States
- 10USDA/ARS Plant Genetics Research Unit, Columbia, MO, United States
Adaptation of soybean cultivars to the photoperiod in which they are grown is critical for optimizing plant yield. However, despite its importance, only the major loci conferring variation in flowering time and maturity of US soybean have been isolated. By contrast, over 200 genes contributing to floral induction in the model organism Arabidopsis thaliana have been described. In this work, putative alleles of a library of soybean orthologs of these Arabidopsis flowering genes were tested for their latitudinal distribution among elite US soybean lines developed in the United States. Furthermore, variants comprising the alleles of genes with significant differences in latitudinal distribution were assessed for amino acid conservation across disparate genera to infer their impact on gene function. From these efforts, several candidate genes from various biological pathways were identified that are likely being exploited toward adaptation of US soybean to various maturity groups.
Introduction
Flowering time is a key trait for maximizing yield potential in many crop species. Extensive research into the genetic mechanisms controlling reproductive timing in the long-day-flowering model organism Arabidopsis thaliana has blazed the trail for understanding the flowering process and has been instrumental in identifying these genes in other species. At least six major pathways coordinate to modulate flowering, including the photoperiod, ambient temperature, vernalization, autonomous, aging, and gibberellins pathways (reviewed in Fornara et al., 2010). Together, members of these pathways integrate environmental and endogenous signals and converge upon key floral integrator genes to promote flowering under optimum conditions (Figure 1).
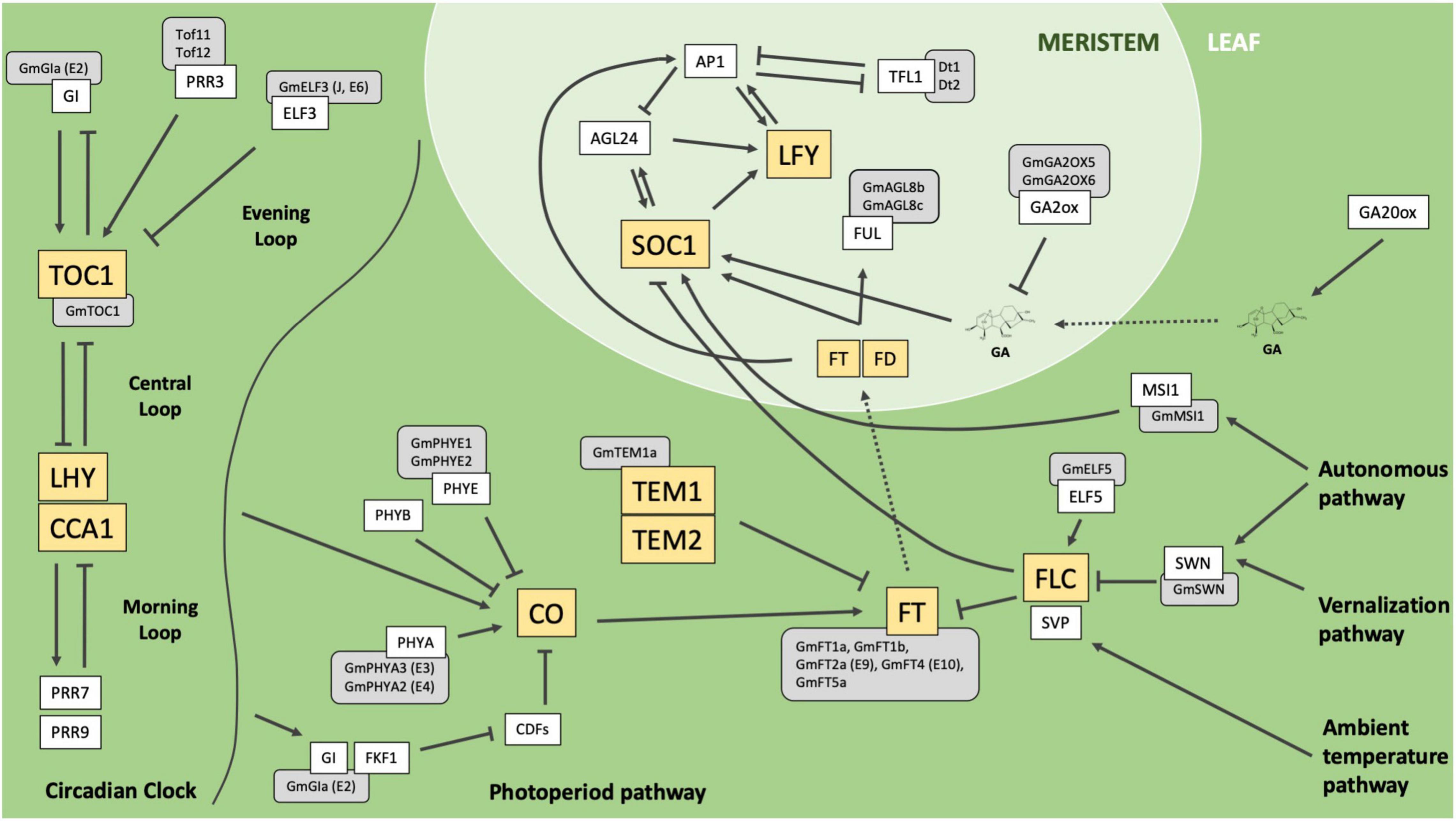
Figure 1. Simplified depiction of characterized pathways and genes which influence flowering in the model organism Arabidopsis thaliana (adapted from Fornara et al., 2010). Sharp boxes represent Arabidopsis genes. Gray rounded boxes represent orthologous genes in soybean. Yellow boxes highlight key integrators or regulators in their respective pathways. Solid arrows indicate positive regulation, T-bars represent negative regulation, dotted arrows represent mobility from leaf to meristem. Arabidopsis genes: Gigantea (GI), Pseudo Response Regulator 3 (PRR3), Early Flowering 3 (ELF3), Timing of CAB Expression 1 (TOC1), Late Elongated Hypocotyl (LHY), Circadian Clock Associated 1 (CCA1), Pseudo Response Regulator 7 (PRR7), Pseudo Response Regulator 9 (PRR9), Flavin-binding, Kelch Repeat, F-Box 1 (FKF1), Cycling DOF Factors (CDFs), Phytochrome A (PHYA), Phytochrome B (PHYB), Phytochrome E (PHYE), Constans (CO), Tempranillo 1 (TEM1), Tempranillo 2 (TEM2), Flowering Locus T (FT), Flowering Locus C (FLC), Early Flowering 5 (ELF5), Short Vegetative Phase (SVP), Swinger (SWN), Multicopy Suppressor of IRA 1 (MSI1), Gibberellin 20-oxidase (GA20ox), Gibberellin 2-oxidase (GA2ox), Fruitful (FUL), Flowering Locus D (FD), Suppressor of Constans 1 (SOC1), Leafy (LFY), Agamous-like 24 (AGL24), Apetala 1 (AP1), Terminal Flower 1 (TFL1). Soybean genes: Gigantea A (GmGIa, E2), Time of Flowering 11 (Tof11), Time of Flowering 12 (Tof12), Early Flowering 3 (GmELF3, J, E6), Timing of CAB Expression 1 (GmTOC1), Phytochrome A3 (GmPHYA3, E3), Phytochrome A2 (GmPHYA2, E4), Phytochrome E1 (PHYE1), Phytochrome E2 (PHYE2), Tempranillo 1a (GmTEM1a), Flowering Locus T 1a (GmFT1a), Flowering Locus T 1b (GmFT1b), Flowering Locus T 2a (GmFT2a, E9), Flowering Locus T 4 (GmFT4, E10), Flowering Locus T 5a (GmFT5a), Early Flowering 5 (GmELF5), Swinger (GmSWN), Multicopy Suppressor of IRA1 (GmMSI1), Gibberellins 2-oxidase 5 (GmGA2OX5), Gibberellins 2-oxidase 6 (GmGA2OX6), Agamous-like 8b (GmAGL8b), Agamous-like 8c (GmAGL8c), Determinant 1 (Dt1), Determinant 2 (Dt2).
The circadian clock complex is composed of three distinct groups of genes, the morning loop, central loop, and evening loop, which regulate one another to maintain an endogenous 24-h oscillation period. LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) promote expression of members of the morning loop, while TIMING OF CAB EXPRESSION 1 (TOC1) regulates members of the evening loop (Alabadì et al., 2001; Gendron et al., 2012). Reciprocal regulation between these three loops ensures proper expression of day/night developmental programs. Photoreceptors in the photoperiod pathway, such as the phytochrome and cryptochrome molecules, perceive different wavelengths of light and, together with output from the circadian clock, affect stabilization of the CONSTANS (CO) protein in the leaf (Valverde et al., 2004). The relative balance of CO, a floral promoter, and the TEMPRANILLO (TEM1, TEM2) repressor genes regulates expression of FLOWERING LOCUS T (FT), a key florigen producing gene, in response to daylength (Castillejo and Pelaz, 2008). FLOWERING LOCUS C (FLC) integrates signaling from both the vernalization and autonomous pathways and, when expressed, functions to delay flowering by repressing key floral integrators SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FT expression (Lee et al., 2000).
Under optimum flowering conditions, FT is expressed in the leaf and produces a mobile florigen that travels to the meristem by way of the phloem. Once in the meristem, FT, in concert with SOC1 and LEAFY (LFY), stimulate the floral meristem transition and activate homeotic genes that give rise to the various floral organs (Parcy et al., 1998; Liu C. et al., 2009). Like FT, gibberellins, a class of hormone involved in broad developmental regulation throughout the plant, are produced in the leaf and travel to the meristem where they promote floral induction by upregulation of SOC1 (Mutasa-Göttgens and Hedden, 2009).
In contrast to Arabidopsis, soybean begins its reproductive period in response to short day photoperiods and lacks a vernalization requirement for proper seed germination. Despite this evolutionary divergence, nearly all the isolated flowering time genes in soybean are orthologs of Arabidopsis genes. GmFT2a (E9) and GmFT5a are two such orthologs of the Arabidopsis FT gene which promote flowering in response to environmental and endogenous signals under appropriate conditions (Takeshima et al., 2016; Zhao et al., 2016). E1, a legume-specific B3-related transcription factor, mediates the photoperiod response to suppress GmFT2a and GmFT5a expression during long days (Xia et al., 2012). Once days shorten past a critical threshold, E1 is suppressed, which derepresses GmFT2a and GmFT5a to allow flowering.
Tof11 and Tof12 are recently isolated paralogs which contributed to soybean domestication (Lu et al., 2020). These genes are orthologs of the Arabidopsis circadian clock gene PSEUDO RESPONSE REGULATOR 3 (PRR3), which function upstream of the central loop oscillator GmLHY. GmPHYA3 (E3) and GmPHYA2 (E4) are orthologs of the Arabidopsis PHYA gene (Liu et al., 2008; Watanabe et al., 2009). These phytochrome molecules perceive red and far-red light, which, in concert with the Tof11, Tof12, and GmLHY, mediate E1 expression to coordinate flowering in response to daylength. Like Tof11 and Tof12, GmGIa (E2) is a circadian clock-associated member of the evening loop in soybean and is a paralog of the Arabidopsis GIGANTEA (GI) gene (Watanabe et al., 2011). GmGIa contributes to flowering in response to photoperiod by delaying expression of GmFT2a under long days. The J locus (E6) is an ortholog of EARLY FLOWERING 3 (ELF3), another circadian clock gene which delays flowering under short day photoperiods and is used to extend the vegetative phase of soybean grown in low latitude production environments (Lu et al., 2017).
Five mutant alleles of the E1 gene have been identified, including alleles containing a frameshift mutation (e1-fs), a missense mutation (e1-as), a retrotransposon insertion (e1-re), a likely regulatory mutation (e1-p), and a complete deletion of the E1 gene (e1-nl) (Tsubokura et al., 2013). A recessive allele of E2 contains a single base substitution resulting in a non-sense mutation (Watanabe et al., 2011). The E3 gene has two variant alleles that differ in their amino acid sequences, relative to the Williams 82 reference: a rare allele containing a non-sense mutation in third exon (e3-mo), and an allele containing a large ∼13 kb deletion after the third exon, resulting in a truncated protein (e3-tr) (Watanabe et al., 2009; Tsubokura et al., 2013). A single mutant allele of the E4 gene contains retrotransposon insertion in its first exon (e4-SORE-1) (Liu et al., 2008).
Soybean varieties are adapted to narrow latitudinal ranges, referred to as Maturity Group (MG), due to their propensity to flower and mature in response to photoperiod. In US soybean production, E1, E2, and E3 confer most of the observed variation in flowering time and maturity; no variation in the E4 gene has been detected in US soybean cultivars (Langewisch et al., 2017). In each case, the dominant allele confers later flowering and maturity. e1-as, a partially functional recessive allele of E1, contains a single missense mutation in its nuclear localization sequence, impairing its ability to enter the nucleus to suppress GmFT2a, and resulting in earlier flowering (Xia et al., 2012). The non-sense mutation in the E2 gene results in a non-functional protein and confers early flowering (Watanabe et al., 2011). The predominant recessive allele of E3 among US soybean, e3-tr, produces a truncated non-functional protein which confers early flowering (Watanabe et al., 2009). However, given the inability of short-read sequencing data to capture large insertions and deletions, the e3-tr mutant allele was not incorportated in this study.
Allelic combinations of these three genes are the major loci conferring adaptation to the various MGs in US soybean varieties (Langewisch et al., 2017). The dominant alleles of all three genes are present in the late flowering cultivars of the south (MG V through MG VIII), whereas accruement of additional recessive alleles confers earlier flowering of cultivars in northern MGs (MG 0–MG IV). Despite this knowledge, our understanding of the loci which confer modest changes in flowering time, such as those within MGs or between adjacent maturity groups, is still lacking. In this work, we utilized resequencing data from 264 elite US soybean lines, converted into a specialized “allele” format, to test soybean orthologs of Arabidopsis flowering genes for differences in latitudinal adaptation between lines differing for alleles. Here, latitude was utilized as a proxy for relative flowering time. For genes with significant p values, weblogos were generated from multiple sequence alignments to determine which mutations occurred in conserved peptide domains and were thus likely to impact protein function. Based on these criteria, we present a set of eight high confidence genes which warrant further investigation as targets for optimizing flowering time and maturity in US soybean cultivars.
Materials and Methods
Curation of Imputed Resequencing Dataset
As part of a parallel effort to catalog the extent of genetic variation in wild and cultivated soybean, our research group previously developed a diversity panel derived from two sets of publicly available resequenced soybean accessions (Zhou et al., 2015; Valliyodan et al., 2021). In this work we used 772 accessions from this diversity panel as a core set of accessions, from which we obtained a total of ∼35.7 M SNP and InDel positions (Škrabišová et al., 2022). We genotyped an additional 518 resequenced accessions at those ∼35.7 Mil positions from other published datasets (Happ et al., 2019; Liu et al., 2020) or from our own skim resequencing efforts (Supplementary Table 1). The set of 65 accessions resequenced by our research group were predominantly elite US soybean lines (ELs) derived from the public university soybean breeding programs in Minnesota, Michigan, Illinois, and Missouri.
Raw reads for publicly available accessions were obtained from NCBI SRA (projects: SRP062245, SRP105183, SRP045129, PRJNA512147) or from the National Genomics Data Center Genome Sequence Archive (project: PRJCA002030). All reads were aligned to the Wm82.a2.v1 Phytozome reference genome using BWA Mem v0.7.17. Variants were called using GATK HaplotypeCaller v4.1.9.0 in “-ERC GVCF” mode. Any identified variant positions which were exclusive to the 518 non-reference panel accessions (i.e., those positions which were not part of the ∼35.7 M SNPs and InDels called from the reference panel) were excluded. Imputation with Beagle v5.2 was performed on the ∼35.7 M positions using the core set of 772 accessions as a reference panel to fill in any missing genotype data derived from regions of poor read coverage or quality. The effect of each of these variants was predicted using the software utility SNPEff v4.3.1t and the Ensembl GTF annotation file for the Wm82.a2.v1 reference genome.1 Variant annotations were restricted to the primary transcript only.
Conversion of Variants to Alleles
The full set of ∼35.7 Mil variant positions was filtered to only include those positions predicted to cause some non-conservative amino acid change in a gene product. This includes all exonal InDels, non-sense mutations, splice site mutations, loss or gain of either a start or stop codon, and non-conservative missense mutations (as defined by groups in Supplementary Table 2). For all accessions, the remaining variant positions in each gene were concatenated to create a putative allele for all ∼55k genes in the genome. The full accession panel was then filtered to obtain only the subset of ELs for which state of origin could be determined; these 264 ELs were then used for subsequent analyses.
Latitude Assignment and Rescaling of 264 Elite US Soybean Lines
For 141 of the 264 resequenced ELs, we were able to obtain days to maturity or relative maturity scores directly from the breeders who developed them (Supplementary Table 3). In such cases, we used those maturity scores to expand the relative latitude values of each EL to the latitudinal range encompassed by the approximate northern and southern border of their respective state. Supplementary Figure 1 illustrates this methodology using 12 resequenced ELs derived from Dr. Brian Diers’ breeding program in Illinois as an example. The northern border of Illinois has a latitude of 42.496369°N, while the southern border has a latitude of 37.231888°N. The earliest maturing EL resequenced from Dr. Diers’ program had a relative maturity (RM) value of 2.5, while the latest maturing EL was assigned an RM of 4.0. The two ELs with RM 2.5 and RM 4.0 were assigned latitude values of 42.496369°N and 37.231888°N, respectively, while the other ten ELs’ latitudes were scaled to this latitudinal range based on their RM values. ELs for which breeders provided days to maturity data, instead of RM values, were treated similarly. In such cases, ELs with the fewest number of days to maturity were assigned a latitude corresponding to the northern border of the state and ELs with the largest number of days to maturity were assigned a latitude corresponding to the southern border of the state. All ELs which matured between the earliest and latest ELs were rescaled to the latitudinal range of the state, according to their respective days to maturity score. ELs which were assigned scaled latitudes based on available maturity information are denoted by “maturity-scaled latitude” in the “LATITUDE_TYPE” column of Supplementary Table 4. ELs for which maturity scores were not available were assigned a latitude value corresponding to the centroidal latitude of the state that the EL originated from (denoted as “centroidal latitude” in the “LATITUDE_TYPE” column of Supplementary Table 4).
E1 and E2 Genotype Assignment of Elite US Soybean Lines From Resequencing Data
E1 and E2 genotypes for all 264 ELs were determined from resequencing results. In addition to the characterized T75R substitution in the E1 gene (Xia et al., 2012), three ELs in our resequencing panel had a frameshift mutation in E1, resulting from a string of adenosine repeats (Supplementary Table 5). Given that InDels which arise from sequence repeats are often artifacts of read alignment error, as well as the low frequency of this mutation among our resequencing panel, only the T75R mutation (06:20207322) was considered when assigning e1-as/E1 genotypes to ELs. Variation at two positions in the E2 gene was present among our resequencing panel: the previously characterized K528* non-sense mutation (10:45310798) leading to the e2 allele (Watanabe et al., 2011), and an isoleucine to valine substitution of amino acid 220 (Supplementary Table 6). As previously stated, only non-conservative mutations were considered when differentiating alleles (as defined by amino acid groups in Supplementary Table 2). As such, E2 genotypes were assigned solely based on the K528* non-sense mutation.
A Curated List of Soybean Orthologs of Arabidopsis Flowering Time Genes Tested for Latitudinal Disparity
Wu et al. (2019) identified 420 soybean orthologs of the 215 Arabidopsis genes involved in flowering from a reciprocal BLASTP query. In this study, we curated an expanded list containing the 420 soybean genes identified by Wu et al., plus an additional 29 orthologs identified from a manual search of the literature (Supplementary Table 7). Genes in this list were excluded from further analysis if (1) they lacked non-conservative mutations, or (2) they lacked at least one alternate allele (i.e., non-reference allele) present in ten or more ELs. These filtering criteria resulted in a final list of 139 genes. These genes were then tested for latitudinal disparity between alleles of the subset of ELs containing the E genotype e1-as/E2. Mean comparisons were carried out using a student t-test (genes with two alleles) or an ANOVA (genes with more than two alleles). The t- or f-statistic representing the 95% confidence interval for each gene was empirically determined by randomization using 1,000 permutations. For genes with more than two alleles, significance letters were obtained from a test for least significant difference. All statistics were conducted using the “stats” package in R v4.0.2 and boxplots for genes with significant p values (p < 0.05*) were generated using ggplot2 v3.3.2.
Weblogo Generation for Genes of Interest
Wm82.a2.v1 peptide sequences for full genes, based on the primary transcript, were pulled from Ensembl using the biomaRt v2.44.4 package. Orthologous sequences were obtained from an NCBI BLASTP query using the command line utility Protein-Protein BLAST v2.10.1. For each returned genus, the orthologous sequence with the highest percent identity was selected, while redundant sequences from each genus were discarded. Multiple sequence alignments for each gene were generated using the msa v1.20.1 R package. Weblogos were generated by clipping the multiple sequence alignment to approximately ten amino acids on either side of each variant, except in cases where variants fell within ten amino acids of the beginning or end of the aligned sequence.
Results
A Curated Set of Resequenced Elite US Soybean Lines
The objective for this work was to identify additional genes that may be contributing to flowering time in US soybean production environments. We performed variant calling and effect annotation on the genome sequence of 264 elite US soybean lines (ELs) from other published works (Zhou et al., 2015; Liu et al., 2020; Valliyodan et al., 2021) and from our own resequencing efforts (Supplementary Table 4). This resulted in a total of ∼11.3 Mil SNPs and InDels. Alleles were defined by the subset of SNPs and InDels which resulted in amino acid changes for each gene. This collection of ELs represents MG’s 0 through VIII and spans a latitudinal range from approximately 27.6°N to 48.9°N. A subset of 31 of these ELs have the maturity genotype E1/E2, 187 ELs have e1-as/E2, and 41 ELs have e1-as/e2 (Supplementary Table 8). The 5 ELs with the maturity genotype E1/e2 were excluded from further analysis (Langewisch et al., 2017).
Soybean Orthologs of Arabidopsis Flowering Genes Tested for Latitudinal Disparity Between Alleles
Alleles for 139 orthologs of Arabidopsis flowering genes (Supplementary Table 9) were tested for significant latitudinal disparity (difference in mean latitudes based on reference or alternate allele(s) of each gene) among the 187 resequenced ELs within the E genotype group e1-as/E2. For genes that were significant (p < 0.05*) for disparities in mean latitude between alleles, a weblogo was created for the variants comprising each allele to determine whether each variant occurred within a conserved sequence domain which is likely important for protein function (Shaner et al., 1993). We identified a set of soybean genes likely playing a role in modulating flowering time in these ELs by taking into consideration latitudinal disparity between alleles, degree of amino acid conservation, and functional annotation of the Arabidopsis orthologs. Only the subset of identified genes, based on the criteria mentioned above, are discussed in further detail here; there were 19 other genes with significant latitudinal disparity that failed to meet our other criteria (Supplementary Figure 2).
E1 and E2 Show Latitudinal Disparity Between Alleles for Elite US Soybean Lines
Mutant alleles of E1 (Glyma.06G207800) and E2 (Glyma.10G221500) confer earlier flowering than their functional counterparts and are exploited to shorten the life cycle of soybean grown in northern US production environments (Watanabe et al., 2011; Xia et al., 2012; Langewisch et al., 2017). As a proof of concept, we analyzed the latitudinal disparity of alleles of E1 and E2 to assess whether latitude was a suitable proxy for flowering time. Among our full resequencing panel, 36 ELs had the functional allele of E1, while 228 ELs had the e1-as missense allele (Supplementary Table 8). A means comparison revealed a significant difference in mean latitude (p < 0.001***), where ELs with the e1-as allele had an average latitude that was 4.6°N higher than those with the E1 allele (Figure 2A). A similar assessment for the E2 gene was conducted using only those ELs containing the e1-as allele of E1. There were 187 ELs in our resequencing panel that had the functional E2 allele, while 41 ELs had the non-sense e2 allele (Supplementary Table 8). Like E1, alleles of E2 showed a significant difference in mean latitude (p < 0.001***), where ELs containing e2 were adapted to 2.5°N higher latitude, on average, than ELs containing E2 (Figure 2B). Our findings concur with previous reports describing the allelic distribution of E1 and E2 among US soybean cultivars and validated our strategy of using latitude as a proxy for relative flowering time and maturity in this study (Langewisch et al., 2017). Furthermore, to avoid the large confounding effect that variation in E1 and E2 would have when assessing the latitudinal disparity of other genes, only the 187 ELs containing the E genotype e1-as/E2 were included in further analysis.
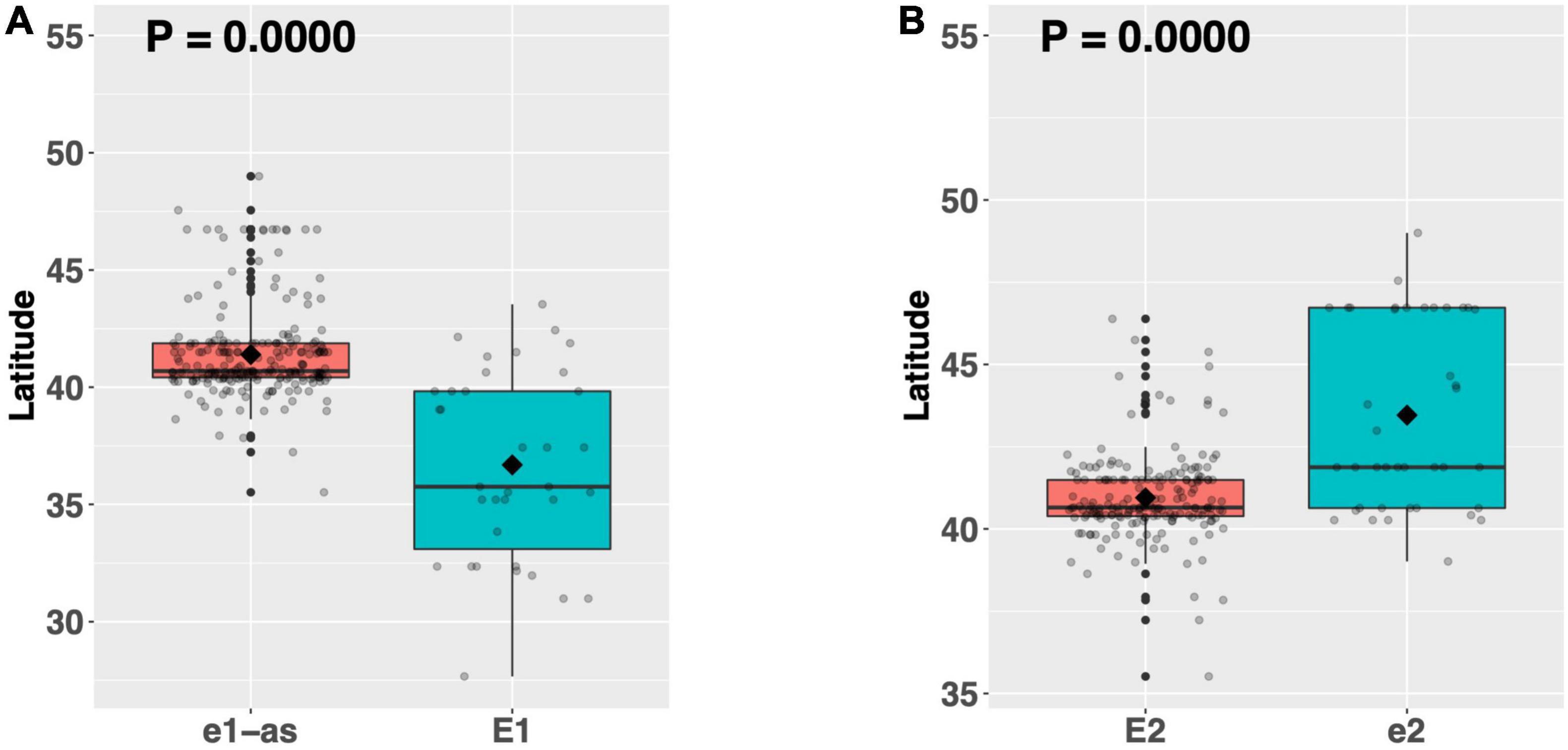
Figure 2. Latitudinal distribution of major alleles of (A) the E1 gene and (B) the E2 gene among 264 resequenced ELs. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparisons were conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). The Williams 82 reference allele is on the left and the box filled with red.
Modulation of Flowering Time by Members of the Autonomous Pathway
The autonomous pathway broadly modulates the plant’s ability to perceive and respond to signaling from other pathways, principally through regulation of FLOWERING LOCUS C (FLC), but also through genes that act in pathways parallel to FLC (Figure 1; reviewed in Simpson, 2004). MULTICOPY SUPPRESSOR OF IRA 1 (MSI1), one such FLC-independent gene, is involved in epigenetic reprogramming of the key floral integrator gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Bouveret et al., 2006). GmMSI1 on chromosome 05 (Glyma.05G131200) was identified as a putative ortholog of the Arabidopsis MSI1 gene (Wu et al., 2019). Among our panel of resequenced ELs, 174 ELs contained the Williams 82 reference allele of GmMSI1, while 20 ELs contained a single methionine to threonine substitution of amino acid 37. We discovered a significant difference (p < 0.05*) in mean latitude between ELs with contrasting alleles of GmMSI1, where ELs with the M37T substitution were adapted to 0.76°N higher, on average, than ELs with the reference allele (Figure 3A). A weblogo generated from 237 genera showed the threonine residue at position 38, as well as the surrounding peptide domain, were highly conserved in this protein and thus likely important for protein function (Figure 3B).
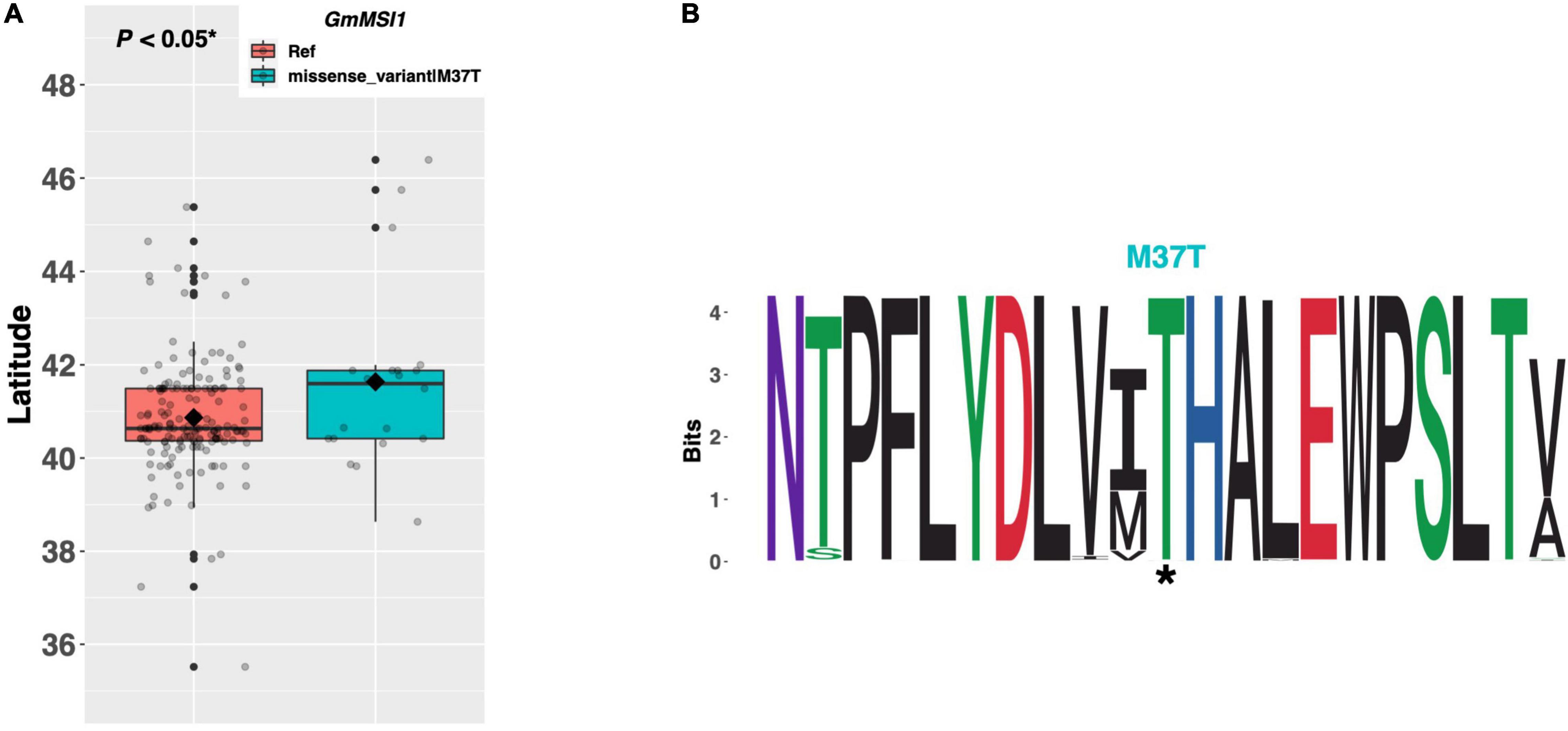
Figure 3. (A) Latitudinal distribution of GmMSI1 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmMSI1.
FLOWERING LOCUS C, a potent repressor of FT and SOC1, is a key floral integrator of endogenous and external signaling from several pathways in Arabidopsis (Figure 1; Lee et al., 2000). Chromatin remodeling of the FLC locus by members of the Polycomb Repressive Complex (PRC), including the histone methyltransferase gene SWINGER (SWN), results in FLC silencing and derepression of FT and SOC1 (Chanvivattana et al., 2004). GmSWN on chromosome 03 (Glyma.03G224300) was identified as a putative ortholog of the Arabidopsis SWN gene (Wu et al., 2019). Our resequencing panel revealed one alternate allele, in addition to the Williams 82 reference allele, containing a threonine to alanine substitution of amino acid 677. There were 37 ELs in the resequencing panel that had the reference allele, while 144 ELs had the T677A missense mutation (Figure 4A). ELs containing the alternate allele of GmSWN were adapted to 0.58°N higher latitude, on average, than ELs containing the reference allele (p < 0.05*), and a weblogo generated from 92 genera showed that the T677A substitution occurred in a region of strong conservation, where alanine was the conserved residue (Figure 4B).
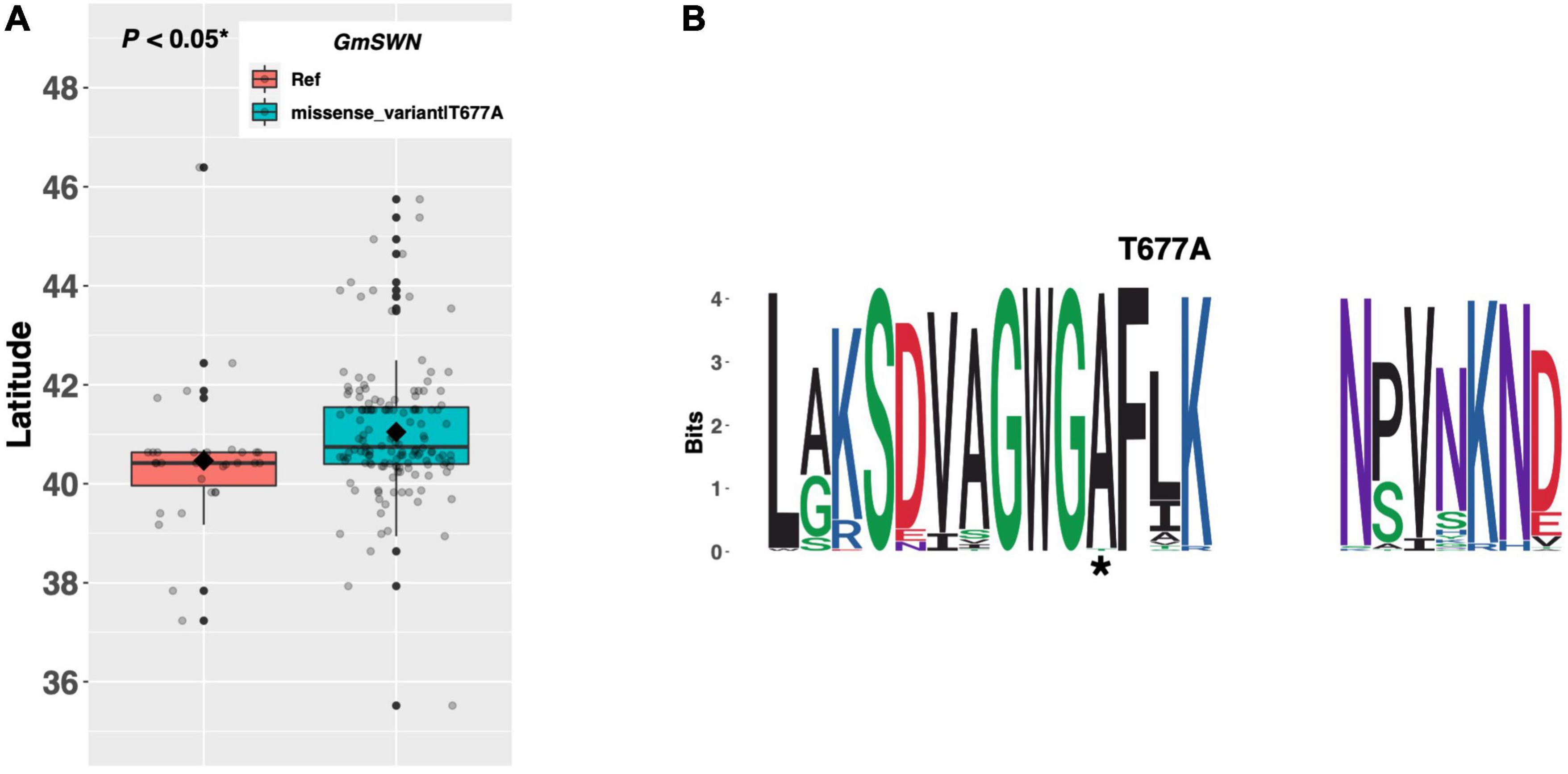
Figure 4. (A) Latitudinal distribution of GmSWN alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparisons were conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisks) in GmSWN.
EARLY FLOWERING 5 (ELF5) is a member of the autonomous pathway in Arabidopsis which has been postulated to post-transcriptionally upregulate FLC to delay flowering (Figure 1; Noh et al., 2004). There are two alternate alleles of the soybean ortholog GmELF5 (Glyma.05G031100) among our resequencing panel, in addition to the Williams 82 reference allele. Both alternate alleles share a glycine to lysine substitution of amino acid 242, a serine to proline substitution of amino acid 202, a serine to leucine substitution of amino acid 282, and an inframe insertion of several amino acids between amino acids 166 and 167 (Figure 5). In addition to these shared mutations, one allele has a mutated splice site and an additional inframe insertion between amino acids 404 and 405, while the other allele has a lysine to asparagine substitution of amino acid 122. A significant latitudinal disparity (p < 0.001***) was observed between the reference allele and the alternate allele containing the splice site mutation, where ELs containing the splice site mutation were adapted to 0.88°N higher latitude than ELs containing the reference allele (Figure 5A). ELs with the alternate allele with the K122N missense mutation were adapted to a similar latitude as the reference allele. A weblogo depicting each of these mutations reveals that the S202P, Q242K, S282L, and the S166_S167 insertion occur in regions of low conservation (Supplementary Figure 3). By contrast, both the P404_P405 insertion and the K122N substitution occur in apparent conserved regions, suggesting that these positions may be important for protein function (Figure 5B).
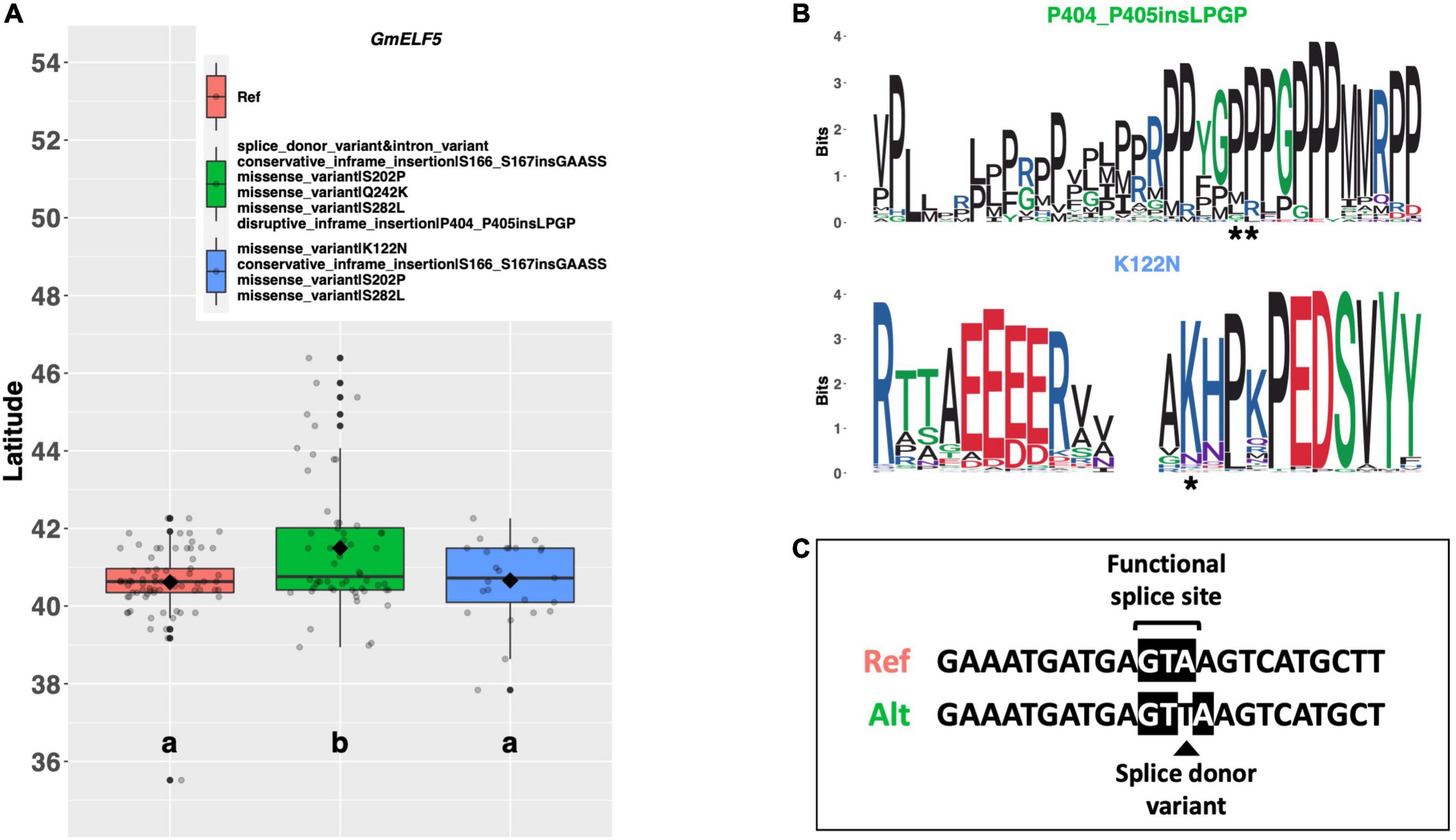
Figure 5. (A) Latitudinal distribution of GmELF5 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using an ANOVA, where the f-statistic representing the 95% confidence interval was empirically derived by randomization. Significance letters were obtained from a test of least significant difference. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding prioritized mutations (asterisk) in GmELF5. (C) Comparison of the splice site sequence at the second exon/intron boundary in the Williams 82 Ref sequence and alternate allele containing the splice donor variant.
The reference allele has an intact splice site, while the allele with the splice donor variant has a single threonine insertion between the second and third nucleotide in the splice site (Figure 5C).
Modulation of Flowering Time by Genes in the Photoperiod and Circadian Clock Pathways
The TEMPRANILLO genes (TEM1/TEM2) are a family of RAV class transcription factors which play a complex role in mediating signaling from several pathways to delay flowering in Arabidopsis (Figure 1; Castillejo and Pelaz, 2008; Osnato et al., 2012). However, despite their key role in Arabidopsis, the effect of TEM orthologs on flowering time in soybean, especially GmTEM1a (Glyma.20G186200), is still underexplored. One alternate allele of GmTEM1a was discovered among the resequenced ELs in the e1-as/E2 genotype group containing a glutamine to proline substitution of amino acid 184 (Figure 6A). ELs with the Q184P substitution were adapted to 0.59°N higher latitude, on average, than ELs with the reference allele (p < 0.05*). A weblogo generated from 124 genera revealed that the proline residue at position 184, as well as the surrounding region, is highly conserved and that the Williams 82 reference line has the non-conserved allele (Figure 6B).
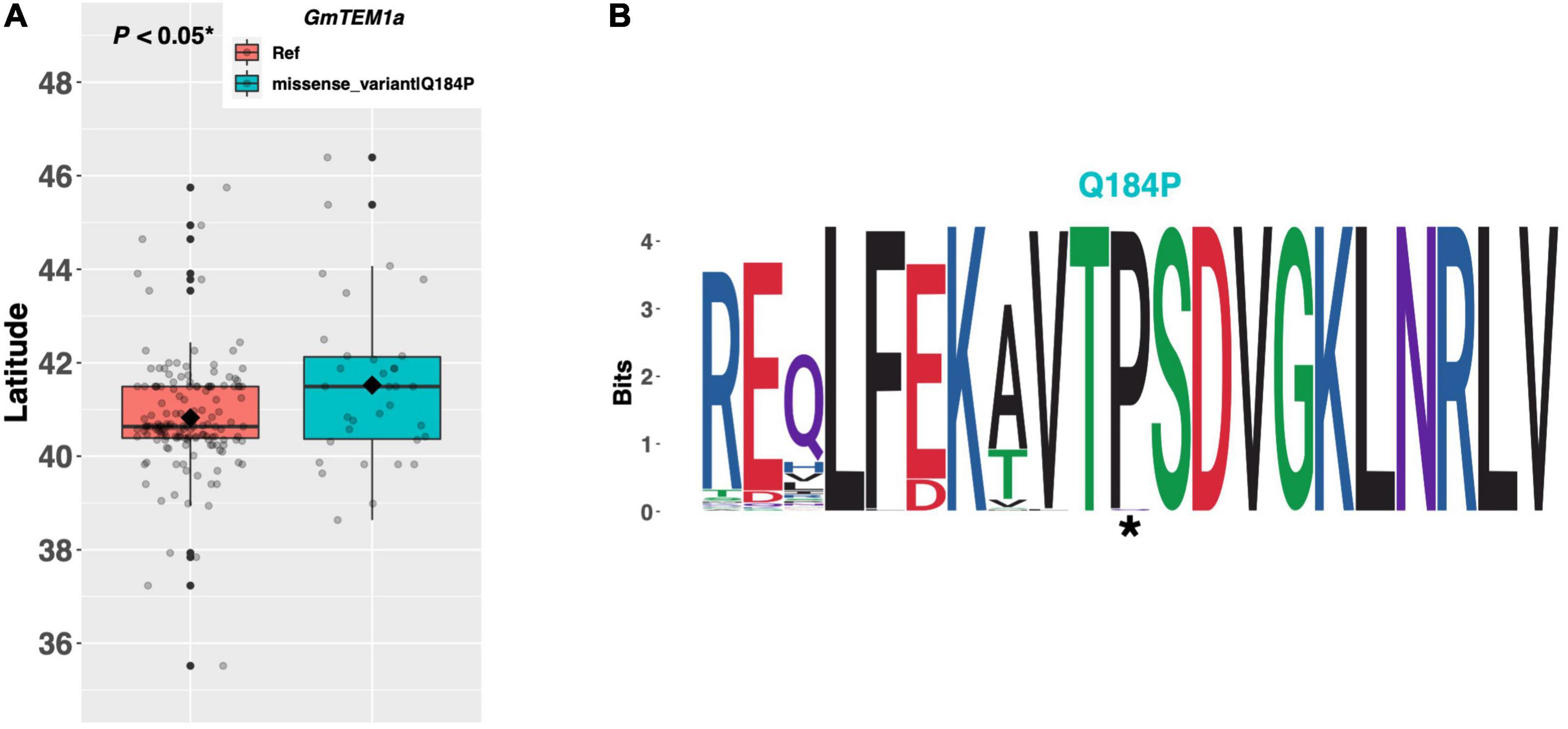
Figure 6. (A) Latitudinal distribution of GmTEM1a alleles among 186 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisks) in GmTEM1a.
In Arabidopsis, PHYTOCHROME E (PHYE) is a photoreceptor molecule which participates broadly in plant developmental responses to light (Figure 1; Devlin et al., 1998). The soybean ortholog GmPHYE1 on chromosome 09 (Glyma.09G088500) has a single alternate allele possessing a histidine to tyrosine substitution of amino acid 708, which is present in 53 ELs in our resequencing panel (Figure 7A). An assessment of latitudinal disparity between the Williams 82 reference allele of GmPHYE1 and the H708Y alternate allele revealed a significant difference in mean latitudes (p < 0.05*), where ELs with the reference allele were adapted 0.58°N higher than ELs with the missense mutation, on average. A weblogo generated from 109 genera showed that the H708Y missense mutation occurred in a conserved protein domain, where the tyrosine residue was conserved and the histidine residue present in Williams 82 was the non-conserved residue (Figure 7B).

Figure 7. (A) Latitudinal distribution of GmPHYE1 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmPHYE1.
TIMING OF CAB EXPRESSION 1 (TOC1) is a core member of the circadian clock complex in Arabidopsis and is responsible for promoting expression of genes in the evening complex (Figure 1; Gendron et al., 2012). The soybean ortholog GmTOC1 (Glyma.06G196200) has a single alternate allele among our resequencing panel, which contains a leucine to serine substitution of amino acid 56 and an isoleucine to serine substitution of amino acid 473 (Figure 8A). An assessment of latitudinal disparity between the Williams 82 reference allele and the alternate allele revealed a significant difference in mean latitude (p < 0.001***), where ELs containing the alternate allele were adapted to 0.64°N higher than ELs with the reference allele (Figure 8A). A weblogo generated from 134 genera showed that the I473S mutation occurred in a region of low conservation, whereas the L56S mutation occurred in a region of high conservation, where the serine residue appeared to be conserved (Figure 8B).
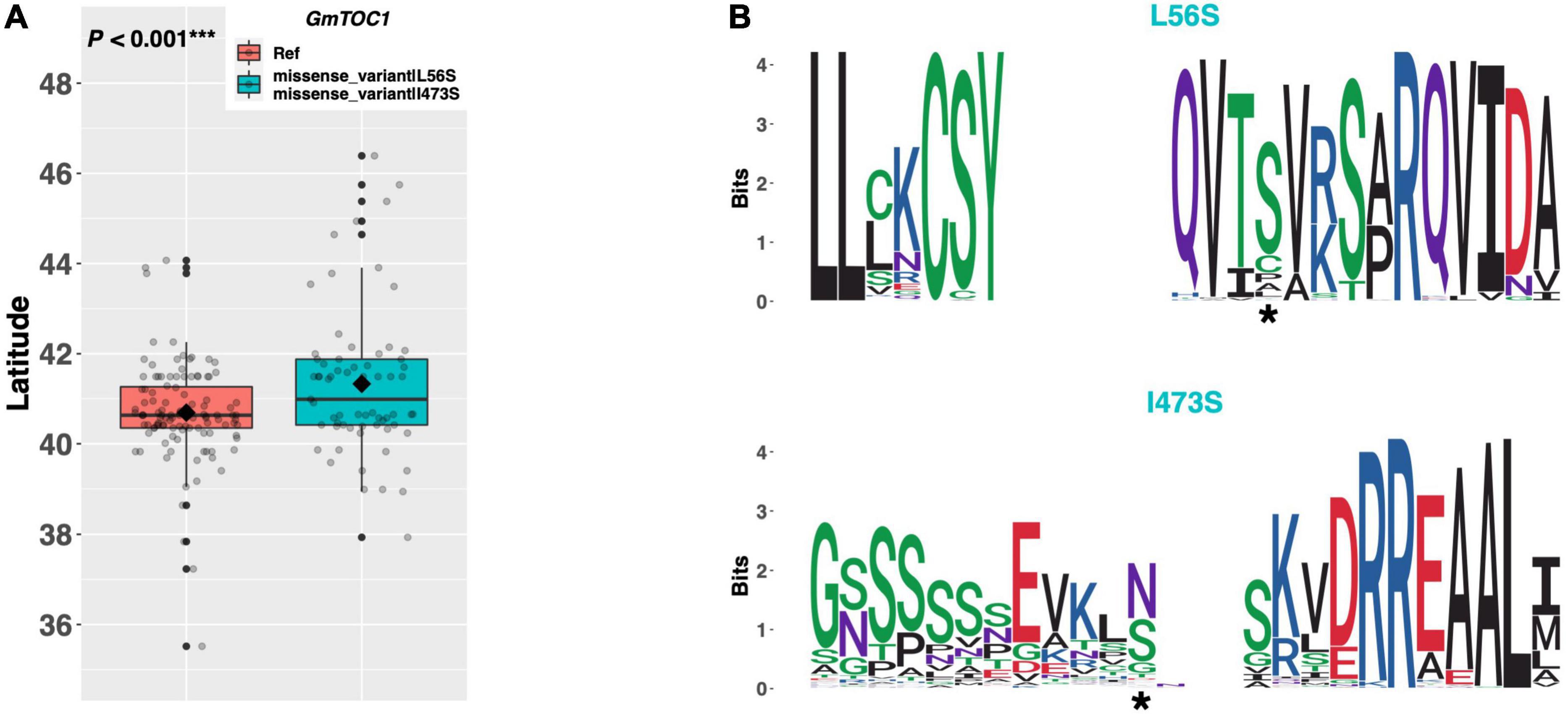
Figure 8. (A) Latitudinal distribution of GmTOC1 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmTOC1.
Modulation of Flowering Time by Genes in the Gibberellin Pathway
Gibberellins are a class of hormone responsible for modulating many developmental programs throughout the plant. GmGA2OX5 and GmGA2OX6 are orthologs of the Arabidopsis GA2OX2 and GA2OX1 genes, respectively, which metabolize gibberellic acid in the apical meristem and delay flowering (Figure 1). GmGA2OX5 (Glyma.13G218200) has a single alternate allele present in 16 ELs in our resequencing panel, which contains a tryptophan to cysteine substitution of amino acid 307 (Figure 9A). We observed a significant difference in mean latitude (p < 0.05*) between ELs containing the Williams 82 reference allele and the alternate allele, where ELs with the reference allele were adapted to 0.67°N higher, on average, than ELs with the alternate allele (Figure 9A). A weblogo generated from 100 genera revealed that the W307C mutation occurred in a region of high conservation, where the tryptophan residue was almost perfectly conserved (Figure 9B).
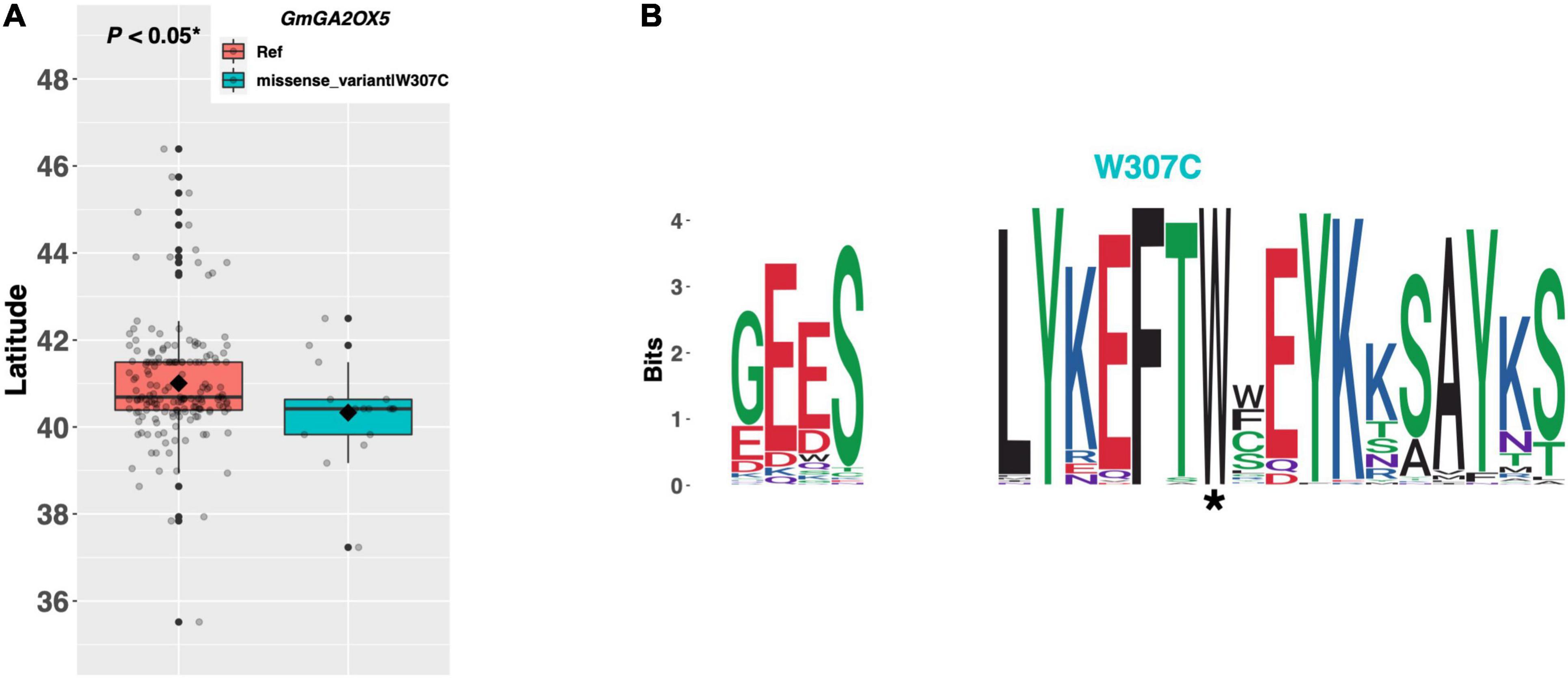
Figure 9. (A) Latitudinal distribution of GmGA2OX5 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmGA2OX5.
Among ELs in our resequencing panel, a single alternate allele of GmGA2OX6 (Glyma.13G259400) existed containing an aspartate to asparagine substitution of amino acid 31, and a proline to alanine substitution of amino acid 182 (Figure 10A). An assessment of latitudinal disparity revealed a significant difference in mean latitude (p < 0.05*) between ELs containing the Williams 82 reference allele and ELs containing the alternate allele, where ELs with the alternate allele were adapted to 0.41°N higher latitude, on average. A weblogo generated from 102 genera revealed that both missense mutations occurred in regions of high amino acid conservation, where the aspartate and proline residues were conserved at positions 31 and 182, respectively (Figure 10B).
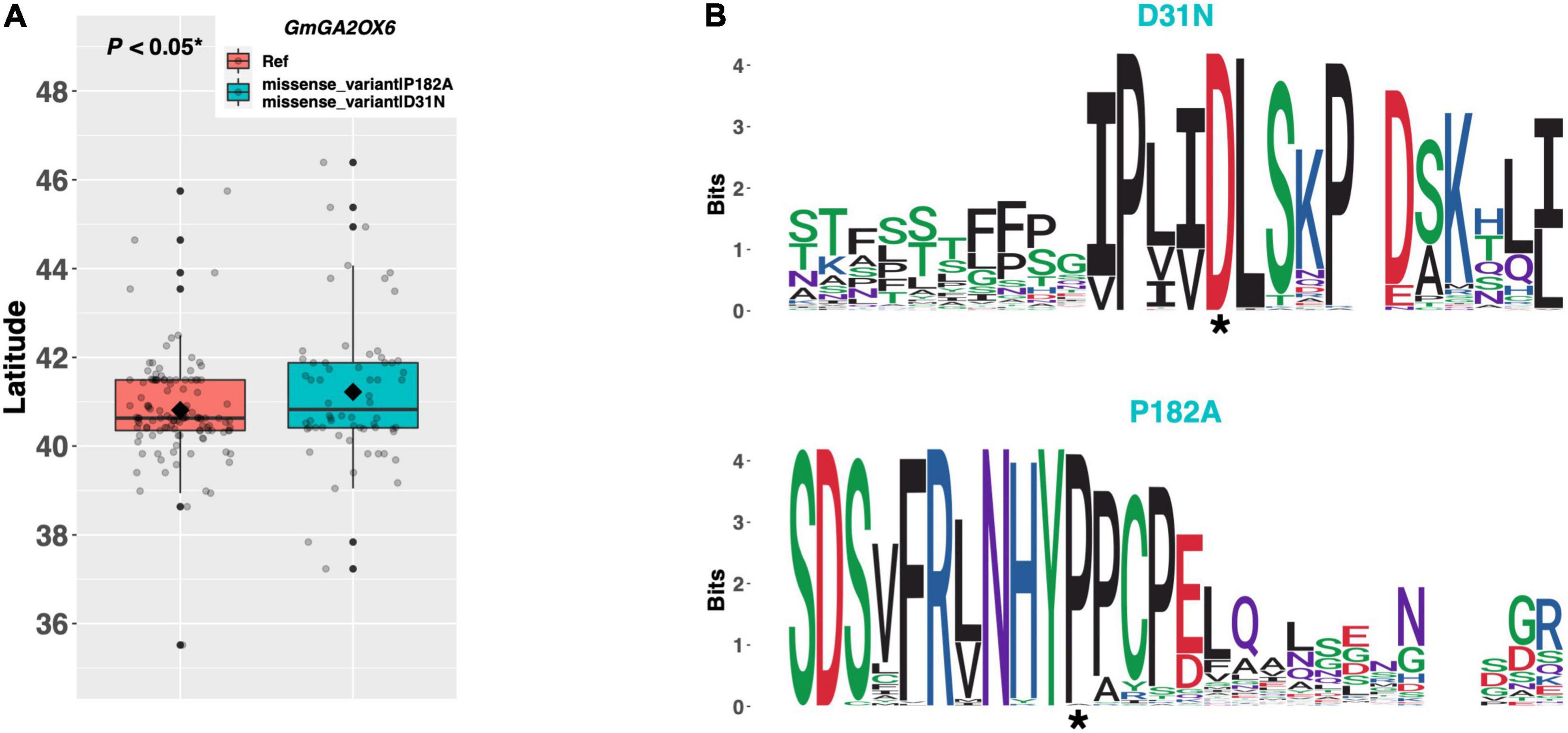
Figure 10. (A) Latitudinal distribution of GmGA2OX6 alleles among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the ELs (see Experimental Procedures). Means comparison was conducted using a student t-test, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele, where “Ref” refers to the Wm82.a2.v1 reference allele. (B) Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmGA2OX6.
Discussion
In this study, we identified an allelic series for soybean orthologs of Arabidopsis flowering time genes from high quality SNPs and InDels derived from resequencing analysis of the genomes of 264 elite US soybean lines (ELs). A total of 139 genes, for which allelic variation was present in high frequency among our resequencing panel, were assessed to determine the likelihood of their utilization to modulate flowering time and maturity in modern US soybean. Eight genes in various biological pathways were identified based on latitudinal disparity between alleles, amino acid conservation of identified mutations, and their function in the model organism A. thaliana. It should be noted that our analysis does not preclude the possibility that enrichment for these alleles in different latitudinal groups could be the consequence of breeder selection for particular alleles of linked genes, rather than selection for the genes identified in this study. However, the degree of sequence conservation, as determined by the weblogos for each variant, suggests these mutations occur in important amino acid domains of their respective proteins and thus likely have an impact on flowering time, regardless of whether they were the intended targets of selection.
Functional Conservation and Divergence of Genes in the Autonomous Pathway
In Arabidopsis, MSI1 promotes flowering via direct upregulation of SOC1 and msi1 mutants resulted in delayed flowering (Bouveret et al., 2006). Our results show that ELs with the putative functional allele of GmMSI1 (i.e., those containing the threonine residue at position 37) are adapted to higher latitudes than ELs with the methionine residue and likely flower and mature earlier. These data suggest that the ability of GmMSI1 to promote floral induction may be conserved in soybean and warrants further investigation as a potential target for optimizing flowering time. As is characteristic of members of chromatin remodeling complexes, mutations in MSI1 pleiotropically affect traits other than flowering time in Arabidopsis, including fertilization and seed development (Köhler et al., 2003). While this seems to suggest that utilizing GmMSI1 to optimize flowering time in soybean may result in developmental abnormalities, the fact that there is significant variation in this gene among ELs, and multiple orthologs of MSI1 exist in soybean, suggests that this gene may have diverged substantially in function from the ancestral state.
In soybean, GmFLC-like acts similarly to FLC by physically interacting with the promoter of GmFT2a to inhibit its expression and delay flowering (Lyu et al., 2020). In Arabidopsis, the SWN gene is expressed in the leaf primordia and apical meristem of young seedlings, as well as in the inflorescence and floral meristems (Chanvivattana et al., 2004). By contrast, GmSWN transcripts were not detected in the Williams 82 reference line (Severin et al., 2010), which contains the putative non-functional allele (i.e., the allele with the threonine residue). The fact that ELs with the putative functional allele (i.e., those with the alanine residue) tend to be adapted to higher latitudes than ELs with the threonine residue, suggests that floral promotion by GmSWN via downregulation of GmFLC-like may be conserved in soybean.
elf5 mutants in Arabidopsis exhibit early flowering in both long and short days at least in part by reduced promotion of the floral repressor FLC (Noh et al., 2004). In soybean cv. Williams 82, GmELF5 is highly expressed in most major tissues throughout the plant (Severin et al., 2010). Our resequencing data indicated several mutations defined two distinct alternate alleles of this gene, in addition to the reference allele. Of particular interest is a mutation in a splice site region defining the second exon/intron boundary, which occurs in one of the alternate alleles. That ELs with this putative null allele are adapted to higher latitudes and likely flower earlier suggests that the function of ELF5 to delay flowering may be conserved in soybean. The second alternate allele contained an asparagine residue in place of a conserved lysine residue at position 122; however, ELs containing this allele seem to be adapted to similar latitudes as ELs with the reference allele. This is at odds with the theory that the function of GmELF5 to delay flowering is conserved, because we would expect ELs with the non-conserved asparagine residue to be adapted to higher latitudes. Further experimentation is needed to determine how GmELF5 and the alleles described herein affect flowering time in soybean.
Functional Conservation and Divergence of Genes in the Photoperiod Pathway
When functional, TEM1/TEM2 compete with the CO protein to inhibit FT expression and delay flowering in Arabidopsis (Castillejo and Pelaz, 2008). Regulatory variation in a light-responsive promoter element of the GmTEM1a paralog, GmTEM1b (also known as GmRAV), was recently shown to promote flowering time and maturity in soybean (Wang et al., 2021), confirming that the native function of GmTEM1b in delaying flowering is conserved between Arabidopsis and soybean. In contrast, ELs with the putative functional allele of GmTEM1a (i.e., those with the proline residue at position 184) are adapted to higher latitudes on average and appear to flower earlier than ELs with the glutamine residue. This finding suggests a possible divergence in function between GmTEM1a and both GmTEM1b and the Arabidopsis TEM1 gene, whereby GmTEM1a may function to promote flowering when functional. Consistent with this idea, Wu et al. showed that, among Clark near-isogenic lines containing contrasting alleles of either E1, E2, or E3, GmTEM1a was differentially expressed under long days in both the E1 and E2 NILs (Wu et al., 2019). By contrast, GmTEM1b was not differentially expressed in any of the three Clark NILs under long days. Taken together, these results suggest that GmTEM1a and GmTEM1b are regulated independently and lends credence to the theory that these genes may have divergent function.
Five phytochrome genes (PHYA to PHYE) exist in Arabidopsis, where PHYA/PHYC and PHYB/PHYD/PHYE form two distinct phylogenetic clades, indicating that the PHYE gene is most closely related to PHYB and PHYD (Mathews, 2006). In Arabidopsis, phye single mutants are identical to wild-type plants but flower early in a phya phyb background, revealing that the function of PHYE overlaps with that of PHYA and PHYB to delay flowering (Devlin et al., 1998). ELs in our resequencing panel which contained the putative functional allele of GmPHYE1 (i.e., those with the conserved tyrosine residue) were adapted to lower latitudes, and thus likely flower later, than those containing the reference allele. GmPHYE1 is expressed in several tissues throughout the Williams 82 reference line, including in the leaves and flowers (Severin et al., 2010). Taken together, these results suggest that the function of PHYE to delay flowering is likely conserved in the soybean ortholog GmPHYE1.
The Arabidopsis TOC1 gene is a core component of the circadian clock involved in reciprocal regulation of genes that stimulate the morning complex (CCA1 and LHY), and impairment of which results in a shortened circadian period and concomitant early flowering (Strayer et al., 2000; Gendron et al., 2012). A novel allele of GmTOC1 (Glyma.06G196200) containing a conserved serine residue at position 56 was identified among ELs in our resequencing panel, and ELs containing this allele were adapted to higher latitudes than ELs with the leucine residue at this position. Liu H. et al. (2009) identified a conserved pseudo-receiver domain near the N terminus of the GmTOC1 paralog on chromosome 04 (Glyma.04G166300) and demonstrated that its expression peaks in the evening, similar to that of TOC1 in Arabidopsis. These data suggest that the function of TOC1 to maintain appropriate circadian period, and thus to delay flowering, appears to be conserved in at least the GmTOC1 gene on chromosome 04. By contrast, those ELs in our resequencing panel with the putative functional allele of the GmTOC1 gene on chromosome 06 (i.e., those with the serine residue) appear to flower earlier, suggesting that there may be divergence in function between GmTOC1 on chromosome 06 and the Arabidopsis TOC1 gene.
Functional Conservation and Divergence of Genes in the Gibberellin Pathway
Biosynthesis of florigenic gibberellic acid (GA) is mediated by GA20OX2 in the leaves of Arabidopsis, the loss of function of which delays flowering modestly under long days and dramatically under short days (Rieu et al., 2008b). By contrast, the GA2OX class of oxidases inactivate GA, and mutants of which result in earlier flowering, especially in short days (Rieu et al., 2008a). However, reports describing the activity of GA reveal that the effects and precise mechanism by which GA influences photoperiod-dependent flowering have significantly diverged between species (reviewed in Mutasa-Göttgens and Hedden (2009)). Our results revealed a single alternate allele in each of two GA2OX orthologs, GmGA2OX5 and GmGA2OX6. ELs in our resequencing panel containing the putative functional allele of GmGA2OX5 (i.e., those with the tryptophan residue at position 307) are adapted to higher latitudes and likely flower earlier than those with the cysteine residue, suggesting that GmGA2OX5 may have diverged in function from its Arabidopsis ortholog, GA2OX. By contrast, ELs containing the putative functional allele of GmGA2OX6 (i.e., those with the aspartate residue at position 31 and the proline residue at position 182) were adapted to lower latitudes than ELs with the putative impaired allele, suggesting that GmGA2OX6 may delay flowering, similar to its Arabidopsis ortholog GA2OX. However, further work is needed to test how each of these genes and the alleles described here affect flowering time in cultivated soybean.
Future Research
The candidate genes identified from this exploratory analysis constitute the preliminary framework needed to further define the genetic architecture of flowering time in US soybean. Given that the latitudinal separation between alleles of these genes is generally smaller than that of E1 and E2, it’s likely that these candidates are minor effect genes, however, further work is needed to validate these as modulators of flowering time and to characterize the magnitude of their effects under natural photoperiods. The fact that eight candidate genes were identified means there are a multitude of allele combinations that may be present in any one accession. When investigating the allele status for the combination those eight genes, accessions with a larger number of the “northern” alleles (the allele with the higher mean latitude) were generally adapted to more northern latitudes, and those with fewer “northern” alleles were adapted to more southern latitudes. Three accessions had the “northern” alleles of all eight candidate genes (PI548540, SS202, and Amcor89), while five accession had just one “northern” allele (PI597382, PI548387, PI548565, Avery, and SA17-15682). Without further research to understand the impact of each identified candidate gene individually, it is difficult to model what is likely to be complex interactions of these genes in relation to photoperiod response. Recent work established a regulatory link between the GmTEM1a ortholog, GmTEM1b (GmRAV), and several gibberellins-associated genes in the modulation of plant height in soybean (Xue et al., 2022). It would be interesting to investigate whether there is a similar link between the GmTEM1a, GmGA2OX5, and GmGA2OX6 genes identified in this study in the control of flowering.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://soykb.org/public_data.php, KB100.
Author Contributions
ND and KB conceived and designed experiments, analyzed data, and wrote the manuscript. YC and TJ developed the resequencing analysis pipelines, generated the combined sequence data files, and provided data quality control. AS, GG, BD, AL, DW, DH, and MH provided germplasm and phenotypes for the analysis. All authors have read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The USDA is an equal opportunity provider and employer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.889066/full#supplementary-material
Supplementary Figure 1 | Example of latitudinal rescaling according to days to maturity or relative maturity scores. Skim resequenced US cultivars (dots) derived from Dr. Brian Diers’ breeding program. Latitude values (left) were scaled according to their Relative Maturity values (right) to fit the approximate latitudinal range of the state of Illinois.
Supplementary Figure 2 | Boxplots of genes with a significant latitudinal disparity between alleles from among 187 resequenced ELs with the maturity genotype e1-as/E2. Latitude of origination was used a proxy for relative flowering time. Latitude values were estimated based on state of origin and, where available, were scaled according to maturity info provided by breeders that developed the cultivars (see Experimental Procedures). Means comparison was conducted using an ANOVA, where the t-statistic representing the 95% confidence interval was empirically derived by randomization. P value refers to significance between alleles with the largest difference in means. Transparent dots represent the latitude of each accession. Boxplots show the mean (diamond), median (solid line), quartile span (box), range (vertical lines), and outliers (solid dots). Inset legend shows the collection of mutations which make up each allele.
Supplementary Figure 3 | Weblogo depicting degree of amino acid conservation of the domain surrounding each mutation (asterisk) in GmELF5.
Footnotes
References
Alabadì, D., Oyama, T., Yanovsky Marcelo, J., Harmon Franklin, G., Más, P., and Kay Steve, A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Bouveret, R., Schönrock, N., Gruissem, W., and Hennig, L. (2006). Regulation of flowering time by Arabidopsis MSI1. Development 133, 1693–1702. doi: 10.1242/dev.02340
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y.-H., Sung, Z. R., et al. (2004). Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Devlin, P. F., Patel, S. R., and Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1487. doi: 10.1105/tpc.10.9.1479
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550–550.e2.
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Happ, M. M., Wang, H., Graef, G. L., and Hyten, D. L. (2019). Generating high density, low cost genotype data in soybean [Glycine max (L.) Merr.]. G3 Genes Genom. Genet. 9, 2153–2160. doi: 10.1534/g3.119.400093
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814. doi: 10.1093/emboj/cdg444
Langewisch, T., Lenis, J., Jiang, G.-L., Wang, D., Pantalone, V., and Bilyeu, K. (2017). The development and use of a molecular model for soybean maturity groups. BMC Plant Biol. 17:91. doi: 10.1186/s12870-017-1040-4
Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J. H., Kim, S.-G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., and Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome a gene. Genetics 180, 995–1007. doi: 10.1534/genetics.108.092742
Liu, C., Xi, W., Shen, L., Tan, C., and Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Liu, H., Wang, H., Gao, P., Xü, J., Xü, T., Wang, J., et al. (2009). Analysis of clock gene homologs using unifoliolates as target organs in soybean (Glycine max). J. Plant Physiol. 166, 278–289. doi: 10.1016/j.jplph.2008.06.003
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-genome of wild and cultivated soybeans. Cell 182, 162–176.e13. doi: 10.1016/j.cell.2020.05.023
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Lyu, J., Cai, Z., Li, Y., Suo, H., Yi, R., Zhang, S., et al. (2020). The floral repressor GmFLC-like is involved in regulating flowering time mediated by low temperature in soybean. Int. J. Mol. Sci. 21:1322. doi: 10.3390/ijms21041322
Mathews, S. (2006). Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15, 3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x
Mutasa-Göttgens, E., and Hedden, P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60, 1979–1989. doi: 10.1093/jxb/erp040
Noh, Y.-S., Bizzell, C. M., Noh, B., Schomburg, F. M., and Amasino, R. M. (2004). EARLY FLOWERING 5 acts as a floral repressor in Arabidopsis. Plant J. 38, 664–672. doi: 10.1111/j.1365-313X.2004.02072.x
Osnato, M., Castillejo, C., Matías-Hernández, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:808. doi: 10.1038/ncomms1810
Parcy, F., Nilsson, O., Busch, M. A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. doi: 10.1038/26903
Rieu, I., Ruiz-Rivero, O., Fernandez-Garcia, N., Griffiths, J., Powers, S. J., Gong, F., et al. (2008b). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504. doi: 10.1111/j.1365-313X.2007.03356.x
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008a). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20, 2420–2436. doi: 10.1105/tpc.108.058818
Severin, A. J., Woody, J. L., Bolon, Y.-T., Joseph, B., Diers, B. W., Farmer, A. D., et al. (2010). RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. doi: 10.1186/1471-2229-10-160
Shaner, M. C., Blair, I. M., and Schneider, T. D. (1993). “Sequence logos: a powerful, yet simple, tool,” in Proceedings of the Twenty-Sixth Hawaii International Conference on System Sciences, (Los Alamitos, CA: IEEE Computer Society Press).
Simpson, G. G. (2004). The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7, 570–574. doi: 10.1016/j.pbi.2004.07.002
Škrabišová, M., Dietz, N., Zeng, S., On Chan, Y., Wang, J., Liu, Y., et al. (2022). A novel synthetic phenotype association study approach reveals the landscape of association for genomic variants and phenotypes. J. Adv. Res. doi: 10.1016/j.jare.2022.04.004
Strayer, C., Oyama, T., Schultz Thomas, F., Raman, R., Somers David, E., Más, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Takeshima, R., Hayashi, T., Zhu, J., Zhao, C., Xu, M., Yamaguchi, N., et al. (2016). A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 67, 5247–5258. doi: 10.1093/jxb/erw283
Tsubokura, Y., Watanabe, S., Xia, Z., Kanamori, H., Yamagata, H., Kaga, A., et al. (2013). Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113, 429–441. doi: 10.1093/aob/mct269
Valliyodan, B., Brown, A. V., Wang, J., Patil, G., Liu, Y., Otyama, P. I., et al. (2021). Genetic variation among 481 diverse soybean accessions, inferred from genomic re-sequencing. Sci. Data 8:50. doi: 10.1038/s41597-021-00834-w
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Wang, Y., Xu, C., Sun, J., Dong, L., Li, M., Liu, Y., et al. (2021). GmRAV confers ecological adaptation through photoperiod control of flowering time and maturity in soybean. Plant Physiol. 187, 361–377. doi: 10.1093/plphys/kiab255
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Watanabe, S., Xia, Z., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Wu, F., Kang, X., Wang, M., Haider, W., Price, W. B., Hajek, B., et al. (2019). Transcriptome-enabled network inference revealed the GmCOL1 feed-forward loop and its roles in photoperiodic flowering of soybean. Front. Plant Sci. 10:1221. doi: 10.3389/fpls.2019.01221
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109, E2155–E2164. doi: 10.1073/pnas.1117982109
Xue, Y., Zhang, Y., Shan, J., Ji, Y., Zhang, X., Li, W., et al. (2022). Growth repressor GmRAV binds to the GmGA3ox promoter to negatively regulate plant height development in soybean. Int. J. Mol. Sci. 23:1721. doi: 10.3390/ijms23031721
Zhao, C., Takeshima, R., Zhu, J., Xu, M., Sato, M., Watanabe, S., et al. (2016). A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16:20. doi: 10.1186/s12870-016-0704-9
Keywords: development, soybean, flowering time, vegetative phase, reproductive phase, genomics, orthologs
Citation: Dietz N, Chan YO, Scaboo A, Graef G, Hyten D, Happ M, Diers B, Lorenz A, Wang D, Joshi T and Bilyeu K (2022) Candidate Genes Modulating Reproductive Timing in Elite US Soybean Lines Identified in Soybean Alleles of Arabidopsis Flowering Orthologs With Divergent Latitude Distribution. Front. Plant Sci. 13:889066. doi: 10.3389/fpls.2022.889066
Received: 03 March 2022; Accepted: 08 April 2022;
Published: 29 April 2022.
Edited by:
Michael Gerard Muszynski, University of Hawai’i at Mānoa, United StatesReviewed by:
Richard Macknight, University of Otago, New ZealandKyuya Harada, Osaka University, Japan
Copyright © 2022 Dietz, Chan, Scaboo, Graef, Hyten, Happ, Diers, Lorenz, Wang, Joshi and Bilyeu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kristin Bilyeu, kristin.bilyeu@usda.gov; bilyeuk@missouri.edu
 Nicholas Dietz1
Nicholas Dietz1 Andrew Scaboo
Andrew Scaboo George Graef
George Graef David Hyten
David Hyten Mary Happ
Mary Happ Brian Diers
Brian Diers Aaron Lorenz
Aaron Lorenz Dechun Wang
Dechun Wang Trupti Joshi
Trupti Joshi Kristin Bilyeu
Kristin Bilyeu