Coastal upwelling enhances abundance of a symbiotic diazotroph (UCYN-A) and its haptophyte host in the Arctic Ocean
- 1Department of Ocean and Earth Sciences, Old Dominion University, Norfolk, VA, United States
- 2Science Philanthropy Alliance, Palo Alto, CA, United States
- 3Woods Hole Oceanographic Institution, Woods Hole, MA, United States
The apparently obligate symbiosis between the diazotroph Candidatus Atelocyanobacterium thalassa (UCYN-A) and its haptophyte host, Braarudosphaera bigelowii, has recently been found to fix dinitrogen (N2) in polar waters at rates (per cell) comparable to those observed in the tropical/subtropical oligotrophic ocean basins. This study presents the novel observation that this symbiosis increased in abundance during a wind-driven upwelling event along the Alaskan Beaufort shelfbreak. As upwelling relaxed, the relative abundance of B. bigelowii among eukaryotic phytoplankton increased most significantly in waters over the upper slope. As the host’s nitrogen demands are believed to be supplied primarily by UCYN-A, this response suggests that upwelling may enhance N2 fixation as displaced coastal waters are advected offshore, potentially extending the duration of upwelling-induced phytoplankton blooms. Given that such events are projected to increase in intensity and number with ocean warming, upwelling-driven N2 fixation as a feedback on climate merits investigation.
Introduction
Marine dinitrogen (N2) fixation, the conversion of N2 gas to ammonia, has historically been ascribed primarily to activity of the filamentous cyanobacterium Trichodesmium in nitrogen (N)-deplete tropical and subtropical waters (e.g., Capone and Carpenter, 1982). However, recent observation of N2 fixation and N2-fixing microbes (diazotrophs) in non-paradigmatic systems has prompted reappraisal of this model (Zehr and Capone, 2020). In particular, the haptophyte-symbiont Candidatus Atelocyanobacterium thalassa (UCYN-A) has received significant attention due to its wide distribution (see Zehr and Capone, 2020) and relatively high diazotrophic activity (e.g., Martínez-Perez et al., 2016). Unlike Trichodesmium, N2 fixation by UCYN-A appears either insensitive to or indirectly stimulated by fixed inorganic N in the environment (e.g., nitrate; Mills et al., 2020), raising questions regarding the responsiveness of this clade to changes in the ocean’s fixed N inventory and obfuscating discussion of the feedback mechanisms involved in N homeostasis. Here, we present data suggesting that UCYN-A and its haptophyte host, Braarudosphaera bigelowii, respond positively and immediately to upwelling of deep, nutrient-rich waters at the shelfbreak of the Beaufort Sea in the western Arctic Ocean, where active N2 fixation has been recently observed at significant rates (Figure 1; Blais et al., 2012; Sipler et al., 2017; Harding et al., 2018).
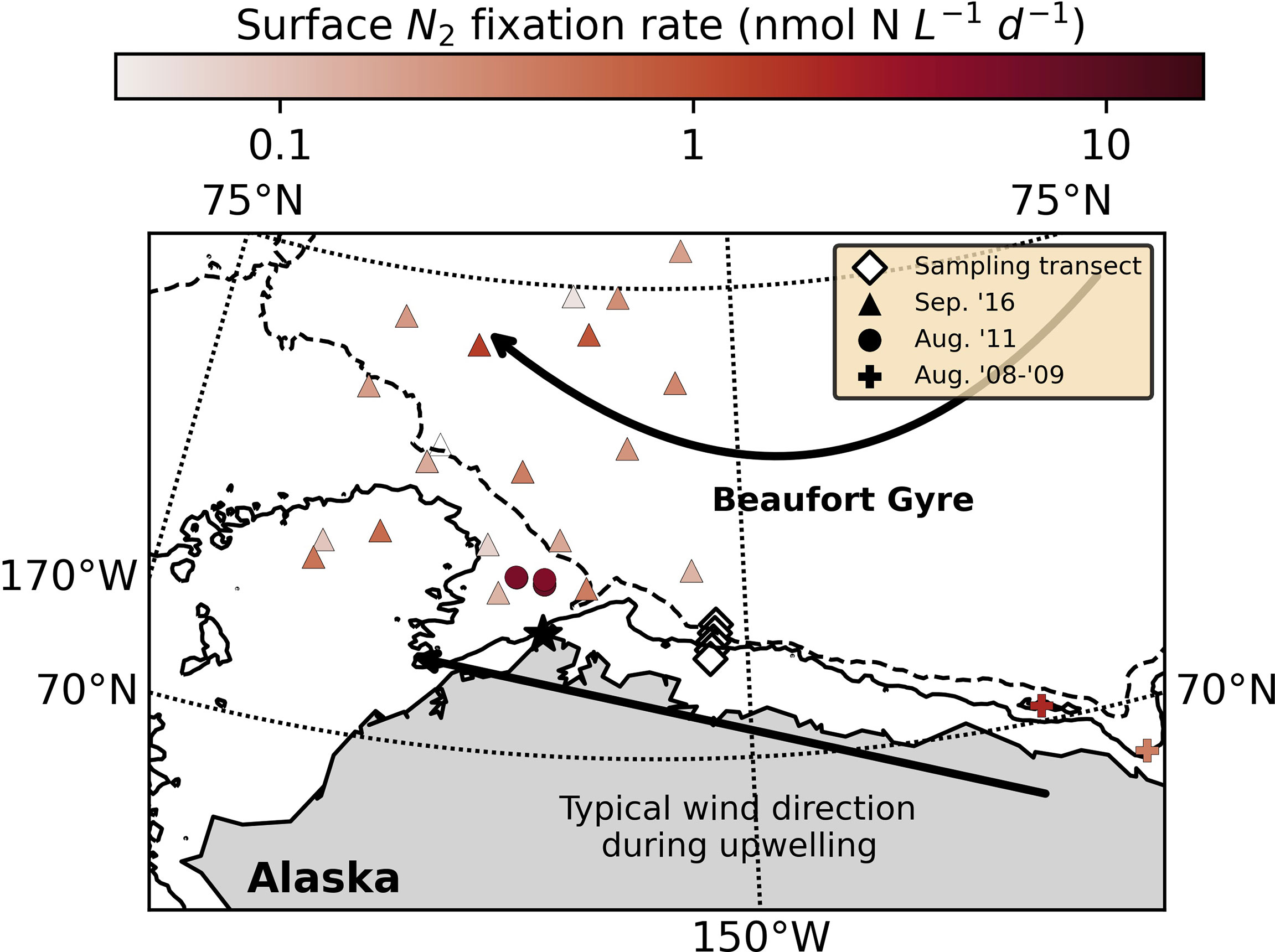
Figure 1 Station locations (diamonds) and N2 fixation rates (not associated with upwelling) measured in August 2008-2009 (crosses; Blais et al., 2012), August 2011 (circles; Sipler et al., 2017) and September 2016 (triangles; Harding et al., 2018). The solid and dashed lines indicate the 50 and 200 m isobaths, respectively (NOAA National Geophysical Data Center 2009; Jakobsson et al., 2012). The Beaufort Gyre is represented schematically by the curved arrow, and the direction of wind that typically induces upwelling is indicated by the straight arrow. Pt. Barrow is labeled with a star.
Materials and methods
Late in the summer of 2017 (August 30 - September 5), the R/V Sikuliaq surveyed waters along the Alaskan Beaufort shelfbreak (Figure 1). The expedition captured different stages of upwelling (see Results and Supplementary Figure 1). Samples and hydrographic profiles were collected along a transect spanning from the mid-shelf to the upper slope. The transect sampled four different time periods: at the start of an upwelling event (August 30-31), during the latter part of the event (September 1-2), shortly after the event concluded (September 3-4), and at the onset of a second, weaker upwelling event (September 5). Along with hydrographic data, we collected surface seawater samples to assess community composition. Upon observing the presence of UCYN-A’s haptophyte host, B. bigelowii, we investigated its abundance and that of two common UCYN-A sublineages, UCYN-A1 and UCYN-A2, found previously in the region (Harding et al., 2018) via targeted quantitative PCR (qPCR).
Hydrographic data collection
Conductivity-temperature-depth (CTD) measurements were made using a Sea-Bird Electronics 911-plus system affixed to a sampling rosette. Nitrate profiles were collected at 7 to 10 stations per transect with an optical nitrate sensor (SUNA V2, Sea-Bird Scientific). To create depth profiles, we aligned the SUNA and CTD data by the recorded time. Water samples from 4-6 depths at 12 representative stations were taken for direct nitrate concentration measurements to calibrate the nitrate sensor. Nitrate concentrations in water samples were measured using an Alpkem RFA continuous flow analyzer following standard colorimetric protocols (Gordon et al., 1993). SUNA nitrate profiles were calibrated by fitting a linear regression to direct measurements from corresponding depths.
Atmospheric fields
To characterize the wind forcing during the expedition, we use the ERA5 reanalysis product from the European Centre for Medium Range Weather Forecasting (ECMWF; Hersbach et al., 2020). The horizontal resolution is 0.25°, and the temporal resolution is 1 hr. The 10 m wind field was used.
DNA sample collection and extraction
Surface water was collected from Niskin bottles mounted to the CTD rosette in dark, 4 L acid-washed (10% HCl) bottles (Nalgene), and promptly filtered (0.22 µm Sterivex™ filters, MilliporeSigma) using a peristaltic pump. Filters were sealed, flash-frozen in liquid N2 and stored at -80°C until DNA extraction. To prepare samples for extraction, the Sterivex™ filters were first removed from their casing in a laminar flow hood using a clean PVC pipe cutter (1% sodium hypochlorite solution soak) to open the case and an autoclaved scalpel to separate the filter. Filters were then transferred in their entirety to sterile microcentrifuge tubes containing AP1 buffer (Qiagen) and beaten for 2 minutes with silica beads. The beads were removed using a QIAshredder spin-column (Qiagen) and DNA was immediately extracted using the DNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. DNA yields following extraction varied from 1267 to 4810 ng per sample.
Community analysis
To investigate eukaryote community diversity, we performed 18S rRNA amplicon sequencing. First, primers derived from the 18S rRNA Illumina Earth Microbiome Project were used to amplify the SSU rRNA 18S V9 marker gene (Stoeck et al., 2010) via polymerase chain reaction (PCR). The products were then gel-purified (GeneJET Gel Extraction Kit, ThermoFisher), subject to a further round of PCR to attach Illumina indices, Mag-Bead purified using AMPure XP beads (Beckman Coulter), and sequenced on Illumina MiSeq Desktop Sequencer at Old Dominion University using a 2×300 bp kit. The DNA sequences were analyzed using the DADA2 pipeline (Callahan et al., 2016); the average number of reads per sample was 66,000. Quality control steps roughly followed defaults of the DADA2 pipeline with a few minor exceptions. Reads without intact primer sequences were discarded from analysis; intact primer sequences were removed from reads using cutadapt (Martin, 2011). Following primer removal, reads were filtered and trimmed. Amplicon sequence variants were identified via BLASTn comparison (Altschul et al., 1990) to an in-house collection of eukaryote 18S rRNA sequences from the National Center for Biotechnology and Information (USA) and SILVA (German Network for Bioinformatics Infrastructure) databases. Only the subset of 18S rRNA reads that were classified as one of the major phytoplankton lineages (dinoflagellates, diatoms, or haptophytes) are presented here. Only B. bigelowii reads were enumerated to the species level.
Diazotroph and host quantification
The abundances of UCYN-A1, UCYN-A2 and B. bigelowii were determined in triplicate via qPCR. Samples were first diluted 1:10 with nuclease-free qPCR-tested ultrapure water (Millipore). Reactions were then prepared by combining 10 µl 2x TaqMan Fast Advanced Master Mix (Applied Biosystems), 4 µl nuclease-free water, 1.6 µl 5 µM custom forward and reverse primers, 0.8 µl 5 µM custom probe, and 2 µl diluted DNA extract (2.5 – 10 ng DNA) for a total reaction volume of 20 µl. These assays targeted nifH sequences (a structural gene associated with the N2 fixation enzyme) specific to two UCYN-A sublineages, UCYN-A1 (Church et al., 2005) and UCYN-A2 (Thompson et al., 2014), and an 18S rRNA sequence associated with a B. bigelowii identified in association with UCYN-A2 (Thompson et al., 2014). Reactions were carried out using a StepOnePlus Real-Time PCR system (Applied Biosystems) following manufacturer’s guidance. Assay efficiencies for this study ranged from 92.4 to 100.4%. Samples, serially-diluted (5 × 10-2–10-9 ng DNA µl-1) synthetic plasmids (GeneWiz), and no-template controls were all run in triplicate; mean values are presented here. The gene concentration range for synthetic plasmid dilution curves ranged from 9.98 × 101-108 gene copies L-1 for B. bigelowii 18S rRNA, and 3.56 × 101-108 gene copies L-1 for UCYN-A1 and UCYN-A2. Effective limits of detection and quantification were calculated as described in Selden et al. (2021) by assuming that the minimum detectable and quantifiable concentrations are 3 and 10 copies reaction-1, respectively (Bustin et al., 2020). These limits varied due to slight differences in sample filtration volume, ranging from 170-220 gene copies L-1 and 575-735 gene copies L-1, respectively.
Statistics
One-way analysis of variance (ANOVA) tests were performed to assess differences among transect occupations in diazotroph and host mean absolute abundances and host relative abundance among all phytoplankton sequences. Calculated test statistics were compared to those computed from 10,000 random permutations of the data to assess significance (Manly, 2007), using an alpha criterion of 0.05. This approach does not require the assumption that the residuals are normally distributed.
Results and discussion
Pre-upwelling conditions
As the upwelling event commenced, the water column in the vicinity of the Beaufort shelfbreak was weakly stratified. Nitrate concentrations in the upper water column were low or undetectable (Figure 2A), and haptophytes represented only a small fraction of the eukaryotic phytoplankton present (≤1%; Supplementary Table 1) on the shelf and slope. The absolute abundance of B. bigelowii was low across the shelf and shelfbreak (mean = 1.44 × 104 gene copies L-1, n = 4 stations; Supplementary Table 1; Figure 2A), but increased at the most seaward station (2.6 × 106 gene copies L-1). UCYN-A1 and UCYN-A2 mirrored this pattern (Supplementary Table 1; Figure 2A). As the abundance of sublineage-specific nifH genes is low compared to the 18S rRNA sequences used to quantify B. bigelowii, UCYN-A1 was only detected at the most offshore station and UCYN-A2 could not be detected at one of the mid-shelf stations (Supplementary Table 1).
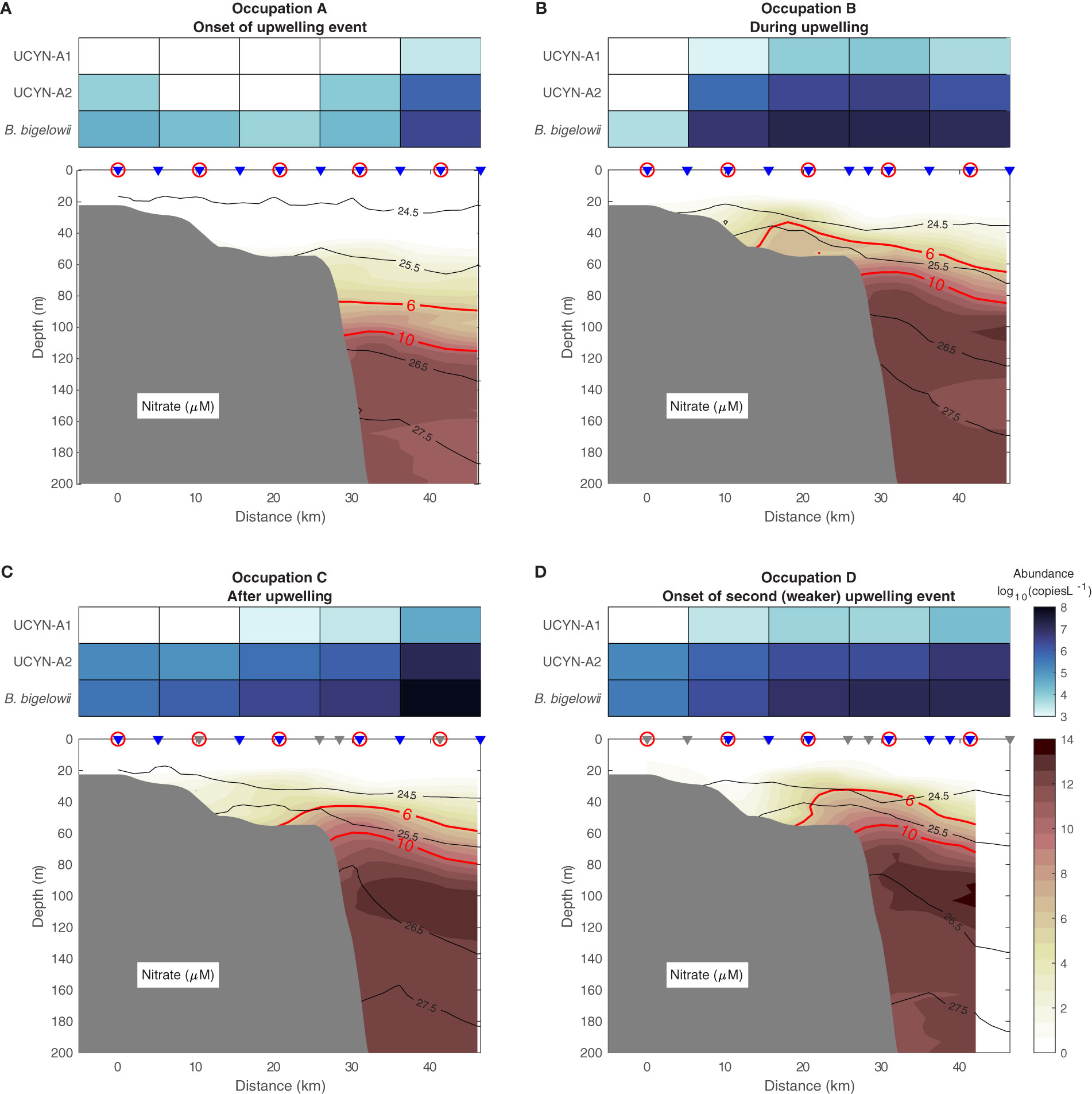
Figure 2 Cross-shelf nitrate concentrations, UCYN-A1 and UCYN-A2 nifH gene abundance, and B. bigelowii 18S rRNA abundance (log10(gene copies L-1)) at the onset of the upwelling event [August 30-31, (A)], during the event [September 1-2, (B)], after the event [September 3-4, (C)], and at the onset of a second, weaker upwelling event [September 5, (D)]. The isopycnals are contoured in black (kg m-3). The contours of 6 and 10 μM nitrate are highlighted in red. The grey triangles are CTD-only stations, the blue triangles are the stations where both CTD and nitrate data were measured, and the stations with gene and rRNA abundance measurements are circled. The bathymetry is from the International Bathymetric Chart of the Arctic Ocean version 3.0 (Jakobsson et al., 2012).
Evidence for upwelling
Upwelling along the Alaskan Beaufort Sea shelf typically occurs when easterly winds exceed 4 m s-1 (Lin et al., 2019). This reverses the normally eastward-flowing shelfbreak jet, and brings water at depth from the basin onto the shelf (Pickart et al., 2011; Schulze and Pickart, 2012). A sub-surface mooring situated just seaward of the shelfbreak, approximately 35 km to the west of the R/V Sikuliaq repeat transect, provided context for the shipboard measurements (Supplementary Figure 1). The along-coast winds became upwelling favorable just as the first transect was occupied (August 30-September 1). Following this, the near-bottom salinity increased and the shelfbreak jet reversed to the west. The second transect (September 1-2) was occupied just past the peak of the event, while the third transect (September 3-4) was carried out after the upwelling had subsided. The final transect (September 5) was done at the start of another event that was considerably weaker.
The first upwelling event caused isopycnal shoaling and drove nitrate-rich waters onto the shelf in a thin stratified layer roughly 20 m above the bottom (Figure 2B). Nitrate concentrations on the shelf decreased as upwelling subsided (Figure 2C), likely due in part to uptake by growing phytoplankton communities (Figure 3) as well as deepening of isopycnals. On the final transect occupation (September 5; Figure 2D), nitrate was again drawn onto the shelf despite the weaker wind event. This is likely because the water column had not fully relaxed from the earlier event, and therefore the previously upwelled water was still located somewhere close to the shelf edge.
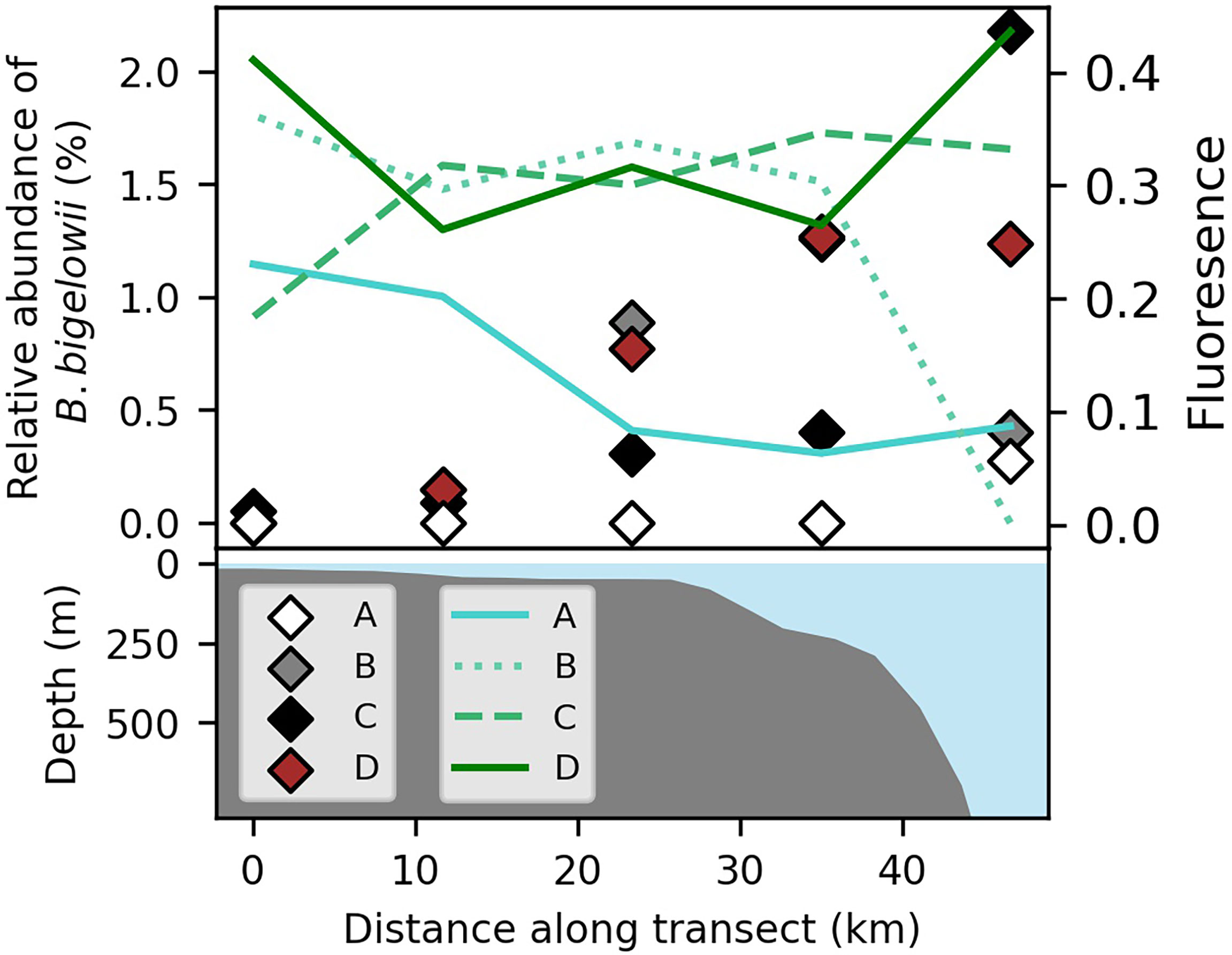
Figure 3 Chlorophyll a relative fluorescence (lines) and relative abundance of B. bigelowii 18s rRNA sequence reads (diamonds) among all eukaryotic phytoplankton (haptophytes, dinoflagellates, diatoms) at the onset of the upwelling event (August 30-31, A), during the event (September 1-2, B), after the event (September 3-4, C), and at the onset of a second, weaker upwelling event (September 5, D; top panel), aligned with bathymetry from the International Bathymetric Chart of the Arctic Ocean version 3.0 (bottom panel; Jakobsson et al., 2012). Blue coloration (bottom panel) indicates water column depth.
Response of UCYN-A and its haptophyte host
Following the onset of upwelling (September 1), chlorophyll-a fluorescence increased (Figure 3) and peaks broadened, suggesting enhanced phytoplankton biomass in the well-lit mixed layer. Concomitantly, we observed a marked increase in the abundance of haptophyte 18S rRNA sequences relative to the sum identified as eukaryotic phytoplankton (diatoms, dinoflagellates, haptophytes) across the transect from a mean of 1.1% to 3.0% (one-way ANOVA, nAug. 30 = 5, nSep. 1 = 5, F = 9.34, p = 0.019), though haptophytes never made up more than 4% of the community at any given station (Supplementary Table 1).
The largest sub-group among haptophyte reads were most closely aligned to sequences from B. bigelowii (>99% identity with coverage of 161/163; Hagino et al., 2013). Prior to upwelling, reads identified as B. bigelowii were only detected in the low-salinity waters (<28 psu) of the offshore end-member (Figure 3; Supplementary Table 1), consistent with prior reports from the region (Shiozaki et al., 2018). The mean relative abundance of B. bigelowii sequences among those of haptophytes, and among those of eukaryotic phytoplankton broadly, increased at all stations during upwelling except at the most coastal station where it remained too low to be observed among 18S rRNA reads (Supplementary Table 1; Figure 3). The highest relative abundances of B. bigelowii were observed at slope stations as upwelling subsided and at the start of the second weaker event, where it represented >1% of eukaryotic phytoplankton (Figure 3) and >40% of haptophyte 18S rRNA reads. While the increase in B. bigelowii as a proportion of the haptophyte population at stations during the onset of upwelling and during peak upwelling were not statistically significant using a standard alpha criterion of 0.050 (one-way ANOVA, nAug. 30 = 5, nSep. 1 = 5, F = 3.92, p = 0.054), likely due to our small sample size, the final occupation (at the beginning of a second, weaker event) bore significantly higher values than the occupation during the onset of upwelling (one-way ANOVA, nAug. 30 = 5, nSep. 5 = 5, F = 6.58, p = 0.041).
The absolute abundance of B. bigelowii 18S rRNA reads correlated with its relative abundance among eukaryotic phytoplankton (Supplementary Figure 2). During upwelling, the absolute abundance of B. bigelowii and both UCYN-A sublineages increased at all stations except the shallowest, where abundance of B. bigelowii and UCYN-A2 decreased (Figure 2; Supplementary Table 1). Nitrate was undetectable in the water column at this station (Figure 2B). B. bigelowii absolute abundance across the entire transect was greater during upwelling than pre-upwelling (one-way ANOVA, nAug. 30 = 5, nSep. 1 = 5, F = 9.16, p = 0.046). Significant differences between pre-upwelling and upwelling conditions were not observed for UCYN-A1 (one-way ANOVA, nAug. 30 = 5, nSep. 2 = 5, F = 4.51, p = 0.094) or UCYN-A2 abundance (one-way ANOVA, nAug. 30 = 5, nSep. 1 = 5, F = 5.03, p = 0.086). As upwelling began to relax, a clear gradient in the abundance of the three organisms appeared with the highest abundances observed at the most offshore station (Figure 2C). By the fourth occupation (at the onset of a weaker upwelling event), abundances of UCYN-A1 (one-way ANOVA, nAug. 30 = 5, nSep. 5 = 5, F = 4.00, p = 0.049) and UCYN-A2 (one-way ANOVA, nAug. 30 = 5, nSep. 5 = 5, F = 3.90, p = 0.009) had significantly increased relative to pre-upwelling conditions.
Given the Eulerian nature of this study, and the physical dynamics of the region, some of the observed enhancement in B. bigelowii and UCYN-A abundances may be attributable to biological patchiness in shelf-waters. However, it is highly unlikely that the observed enhancement resulted solely from alongshore transport of shelf waters because B. bigelowii reads were too rare to be detected among 18S rRNA amplicon sequences collected on the shelf to the east of the study site at the onset of upwelling (Einarsson, 2021). Indeed, they were only detected to the east in waters seaward of the shelf-break, as was the case for the transect described in this study.
As a thought experiment, we calculated doubling times for sequence abundance at each station (Supplementary Table 1). These values were largely reasonable when compared to doubling times for cells (1+ days)—a comparison which assumes that the rate of DNA replication is equal to the rate of cell division. However, B. bigelowii 18S rRNA doubling times at the three intermediate shelf stations during the second occupation (upwelling; September 1-2), and the most coastal station at third occupation (relaxation; September 3-4), showed extreme rates of increase which were unrealistic when compared to doubling times for cells. This trend can be explained by changes in ploidy. Many eukaryotes undergo cyclic changes in ploidy related to their growth cycles and cell size (Parfrey et al., 2008). High levels of DNA are observed in large vegetative cells (Kondorosi et al., 2000). Increases in haptophyte ploidy are thus consistent with the hypothesis that upwelling relieved growth limitation of the diazotroph-haptophyte symbiosis. We note that UCYN-A also displayed rapid DNA doubling times at certain stations; recent work indicates that some cyanobacteria do contain multiple copies of their genome and that this number can greatly vary at different growth stages (Griese et al., 2011; Oliverio and Katz, 2014). As discussed above, community patchiness, too, represents a confounding variable in these calculations.
Throughout the study, the abundances of B. bigelowii 18S rRNA and UCYN-A2 nifH sequences remained tightly correlated at a ratio of ~5 18S rRNA:1 nifH gene copy (linear regression, R2 = 0.966, p < 10-11). In conjunction with recent work from Mills et al. (2020), who found that B. bigelowii relies on diazotroph-derived N to meet its N demand even when other forms of N are available, this finding suggests that UCYN-A was continuously fixing N for its host. It is thus reasonable to expect that volumetric N2 fixation rates increase with abundance of the symbiosis.
Taken together, the results presented here indicate that upwelling favored growth of UCYN-A and its host, B. bigelowii, in surface waters across the Beaufort shelfbreak. In so doing, the upwelling event likely drove increased N2 fixation as well. We propose two potential explanatory mechanisms: (1) the growth of UCYN-A and its symbiont were indirectly stimulated by nitrate (e.g., by increased availability of plankton exudates) as reported from the California Current System by Mills et al. (2020), and/or (2) the UCYN-A/haptophyte symbiosis proliferated because non-diazotrophic phytoplankton drew down nitrate in advance of other upwelled nutrients and the community became N-limited while phosphate and iron remained available, creating a niche for the diazotrophic symbiosis. As samples for DNA analysis were only collected from surface waters, where nitrate concentrations were always low due to rapid biological utilization, and because samples for phosphate determination were not collected, we unfortunately cannot speculate as to mechanism at this time.
Regardless of the exact mechanism, the response of the UCYN-A/haptophyte symbiosis to upwelling observed here may be occurring in upwelling systems globally. Subramaniam et al. (2013) observed enhanced N2 fixation rates following upwelling in the equatorial Atlantic where UCYN-A is known to be highly active (Martínez-Perez et al., 2016). Additionally, Sohm et al. (2011) observed high N2 fixation rates in the Benguela upwelling system. Though they assessed the abundance of a range of diazotrophs, these authors were unable to identify the diazotroph responsible. However, the primers now commonly used for UCYN-A2 and its host had not yet been developed at the time of their study.
Finally, these results suggest that Arctic Ocean N2 fixation may be highly variable as local diazotrophs respond to dynamic physical conditions. This variability is likely not reflected in the only published estimate of N2 fixation on the Arctic shelf (3.5 Tg N y-1 or ~2.7% of global N2 fixation; Sipler et al., 2017), which is based on relatively few data points with low spatial and temporal coverage. Considering upwelling-driven N2 fixation could thus significantly augment basin-wide fixed-N input estimates in the Arctic, as well as in other upwelling systems.
Conclusions
This study presents the novel observation that the abundance of the diazotroph UCYN-A and its haptophyte host increases during and after upwelling at the Beaufort Sea shelfbreak. As UCYN-A appears to be the primary N source for its host (Mills et al., 2020), it is reasonable to infer that N2 fixation rates would be similarly enhanced, though this inference requires further investigation. In polar regions (Harding et al., 2018; Shiozaki et al., 2020), UCYN-A fixes N2 at per-cell rates comparable to those observed in the (sub)tropical North Atlantic (Krupke et al., 2015; Martínez-Perez et al., 2016), where it is responsible for ~20% of N2 fixation (Martínez-Perez et al., 2016). Even a moderate enhancement in rates associated with upwelling may be meaningful for the N budget at local to regional scales. As N2 fixation rates are difficult to quantify, direct measurements typically exhibit poor coverage through space and time and thus rarely resolve short-lived physical features adequately. Our findings suggest that N2 fixation may be underestimated not only in the Arctic, but in other upwelling coastal systems where UCYN-A thrives. More widespread application of high-resolution approaches to studying diazotrophy (Benavides and Robidart, 2020) are necessary to discern the impact of ocean physics on N2 fixation. Doing so may be particularly important in the Arctic Ocean, where the number and intensity of upwelling events is increasing (Pickart et al., 2013)—a trend that may be exacerbated by warming-induced ice retreat. Previous work from the region (Shiozaki et al., 2018) has found that, where UCYN-A2 is present, ~2% of new production (i.e., that driven by upwelled nitrate or N2 fixation) derives N from N2 fixation. If diazotroph-derived N augments upwelling-driven primary productivity, then it may represent a negative climate feedback by enhancing the biological drawdown of atmospheric carbon dioxide. The extent to which upwelling-driven N2 fixation facilitates exhaustion of upwelled phosphorus, and whether the carbon fixed as a result is effectively sequestered, thus merit inquiry.
Data availability statement
Sequencing data can be found in the NCBI database (accession #PRJNA743005): https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743005. All other data are available in supplemental materials.
Author contributions
CRS and SVE co-led the manuscript effort and contributed equally. CRS and PDC originated the research question and designed the analytical approach. KEL and CJA designed the field survey and collected samples and hydrographic data. SVE and KEC conducted DNA analyses. SVE processed hydrographic data and generated figures. CRS, SVE, KEC, and PDC contributed to writing the paper. RSP and PL helped with the interpretation of the environmental data. All authors reviewed and approved of the final manuscript.
Funding
Funding for this work was provided by awards from the Jeffress Trust Awards Program in Interdisciplinary Research to PDC, the Jacques S. Zaneveld endowed scholarship to SVE, and the Weston Howland Jr. Postdoctoral Scholarship and WHOI Access to Sea Fund to KEL. Fieldwork was supported by National Science Foundation grants PLR-1603941 to CJA and Joel Llopiz (Woods Hole Oceanographic Institution), PLR-1603120 to Stephen Okkonen (University of Alaska Fairbanks), and PLR-1603259 to Kathleen M. Stafford (University of Washington). Additional analysis was supported by National Science Foundation grant OPP-1733564 and National Oceanic and Atmospheric Administration grant NA19OAR4320074.
Acknowledgmnts
We thank the captain, crew, and marine science technicians of the R/V Sikuliaq for facilitating sample collection, Kim Powell (Old Dominion University) for assisting in DNA/RNA sample processing, Amala Mahadevan (Woods Hole Oceanographic Institution) for the use of her SUNA sensor, Steve Okkonen (University of Alaska Fairbanks) for help identifying the physical dynamics of the upwelling event, and two reviewers for their sound suggestions. Post-cruise analysis of nitrate samples used to calibrate SUNA sensor was performed at Oregon State University through in-kind contribution by Laurie Juranek.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.877562/full#supplementary-material
References
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Molec. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Benavides M., Robidart J. (2020). Bridging the spatiotemporal gap in diazotroph activity and diversity with high-resolution measurements. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.568876
Blais M., Tremblay J. E., Jungblut A. D., Gagnon J., Martin J., Thaler M., et al. (2012). Nitrogen fixation and identification of potential diazotrophs in the Canadian Arctic. Global Biogeochem. Cy. 26, GB3022. doi: 10.1029/2011gb004096
Bustin S. A., Mueller R., Nolan T. (2020). “Parameters for successful PCR primer design,” in Quantitative real-time PCR: Methods and protocols. methods in molecular biology. Eds. Biassoni R., Raso A. (Humana, New York, NY: Springer Science and Business Media, LLC, part of Springer Nature).
Callahan B. J., McMurdie P. J., Rosen M., Han A., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Meth. 13, 581–583. doi: 10.1038/nmeth.3869
Capone D. G., Carpenter E. J. (1982). Nitrogen fixation in the marine environment. Science 217, 1140–1142. doi: 10.1126/science.217.4565.1140
Church M. J., Jenkins B. D., Karl D. M., Zehr J. (2005). Vertical distributions of nitrogen-fixing phylotypes at stn ALOHA in the oligotrophic north pacific ocean. Aquat. Microb. Ecol. 38, 3–14. doi: 10.3354/ame038003
Einarsson S. (2021). Phytoplankton community response to upwelling events: Distribution and abundance investigated using genomic methods (Norfolk (VA: Old Dominion University).
Gordon L. I., Jennings J. C. Jr., Ross A. A., Krest J. M. (1993). A suggested protocol for continuous flow automated analysis of seawater nutrients (phosphate, nitrate, nitrite and silicic acid) in the WOCE hydrographic program and the joint global ocean fluxes study (WOCE Hydrographic Program Office, Methods Manual WHPO), 1–52.
Griese M., Lange C., Soppa J. (2011). Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323 (2), 124–131. doi: 10.1111/j.1574-6968.2011.02368.x
Hagino K., Onuma R., Kawachi M., Horiguchi T. (2013). Discovery of an endosymbiotic nitrogen-fixing UCYN-a in Braarudosphaera bigelowii (Prymnesiophyceae). PloS One 8 (12), e81749. doi: 10.1371/journal.pone.0081749
Harding K., Turk-Kubo K., Sipler R., Mills M., Bronk D., Zehr J. (2018). Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic ocean. P. Natl. Acad. Sci. U.S.A. 115, 13371–13375. doi: 10.1073/pnas.1813658115
Hersbach H., Bell B., Berrisford P., Hirahara S., Horányi A., Muñoz-Sabater J., et al. (2020). The ERA5 global reanalysis. Q. J. R. Meteorol. Soc. 146 (730), 1999–2049. doi: 10.1002/qj.3803
Jakobsson M., Mayer L. A., Coakley B., Dowdeswell J. A., Forbes S., Fridman B., et al (2012). The international bathymetric chart of the Arctic ocean (IBCAO) version 3.0. Geophys. Res. Lett. 39 (12), L12609. doi: 10.1029/2012GL052219
Kondorosi E., Roudier F., Gendreau E. (2000). Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3 (6), 488–492. doi: 10.1016/s1369-5266(00)00118-7
Krupke A., Mohr W., LaRoche J., Fuchs B., Amann R., Kuypers M. (2015). The effects of nutrients on carbon and nitrogen fixation by the UCYN-a haptophyte symbiosis. ISME J. 9, 1635–1647. doi: 10.1038/ismej.2014.253
Lin P., Pickart R. S., Moore G. W. K., Spall M. A., Hu J. (2019). Characteristics and dynamics of wind-driven upwelling in the alaskan Beaufort Sea based on six years of mooring data. Deep-Sea Res. Pt II 162, 79–92. doi: 10.1016/j.dsr2.2018.01.002
Manly B. F. (2007). Randomization, bootstrap and Monte Carlo methods in biology (New York, NY: CRC Press).
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Martínez-Perez C., Mohr W., Löscher C., Dekaezemacker J., Littmann S., Yilmaz P., et al. (2016). The small unicellular diazotrophic symbiont, UCYN-a, is a key player in the marine nitrogen cycle. Nat. Microbiol. 1, 16163. doi: 10.1038/NMICROBIOL.2016.163
Mills M. M., Turk-Kubo K., van Dijken G., Henke B., Harding K., Wilson S., et al. (2020). Unusual marine cyanobacteria/haptophyte symbiosis relies on N2 fixation even in n-rich environments. ISME. J. 14, 2395–2406. doi: 10.1038/s41396-020-0691-6
Oliverio A., Katz L. (2014). The dynamic nature of genomes across the tree of life. Genome Biol. Evol. 6 (3), 482–488. doi: 10.1093/gbe/evu024
Parfrey L., Lahr D., Katz L. (2008). The dynamic nature of eukaryote genomes. Molec. Biol. Evol. 25 (4), 787–794. doi: 10.1093/molbev/msn032
Pickart R. S., Schulze L. M., Moore G. W. K., Charette M. A., Arrigo K. R., van Dijken G., et al. (2013). Long-term trends of upwelling and impacts on primary productivity in the alaskan Beaufort Sea. Deep-Sea Res. Pt I: Oceanographic Research Papers 79, 106–121. doi: 10.1016/j.dsr.2013.05.003
Pickart R. S., Spall M. A., Moore G. W. K., Weingartner T. J., Woodgate R. A., Aagaard K., et al. (2011). Upwelling in the alaskan Beaufort Sea: Atmospheric forcing and local versus non-local response. Prog. Oceanogr. 88 (1-4), 78–100. doi: 10.1016/j.pocean.2010.11.005
Schulze L. M., Pickart R. S. (2012). Seasonal variation of upwelling in the alaskan Beaufort Sea: Impact of sea ice cover. J. Geophys. Res-Oceans 117, C06022. doi: 10.1029/2012JC007985
Selden C., Chappell P. D., Clayton S., Macías-Tapia A., Bernhardt P., Mulholland M. R. (2021). A coastal N2 fixation hotspot at the cape hatteras front: Elucidating spatial heterogeneity in diazotroph activity via supervised machine learning. Limnol. Oceanogr. 66:5, 1832–1849. doi: 10.1002/lno.11727
Shiozaki T., Fujiwara A., Ijichi M., Harada N., Nishino S., Nishi S., et al. (2018). Diazotroph community structure and the role of nitrogen fixation in the nitrogen cycle in the chukchi Sea (western Arctic ocean). Limnol. Oceaogr. 63, 2191–2205. doi: 10.1002/lno.10933
Shiozaki T., Fujiwara A., Inomura K., Hirose Y., Hashihama F., Harada N. (2020). Biological nitrogen fixation detected under Antarctic sea ice. Nat. Geosci. 13, 729–732. doi: 10.1038/s41561-020-00651-7
Sipler R. E., Gong D., Baer S. E., Sanderson M. P., Roberts Q. N., Mulholland M. R., et al. (2017). Preliminary estimates of the contribution of Arctic nitrogen fixation to the global nitrogen budget. Limnol. Oceanogr. Lett. 2, 159–166. doi: 10.1002/lol2.10046
Sohm J., Hilton J. A., Noble A. E., Zehr J. P., Saito M. A., Webb E. A. (2011). Nitrogen fixation in the south Atlantic gyre and the benguela upwelling system. Geophys. Res. Lett. 33, L16608. doi: 10.1029/2011GL048315
Stoeck T., Bass D., Nebel M., Christen R., Jones M., B. H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molec. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Subramaniam A., Mahaffey C., Johns W., Mahowald N. (2013). Equatorial upwelling enhances nitrogen fixation in the Atlantic ocean. Geophys. Res. Lett. 40, 1766–1771. doi: 10.1002/grl.50250
Thompson A., Carter B. J., Turk-Kubo K., Malfatti F., Azam F., Zehr J. P. (2014). Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-a and its prymnesiophyte host. Environ. Microbiol. 16, 3238–3249. doi: 10.1111/1462-2920.12490
Keywords: N2 fixation, diazotroph, UCYN-A, Braarudosphaera bigelowii, Arctic Ocean, upwelling, shelfbreak, nitrogen
Citation: Selden CR, Einarsson SV, Lowry KE, Crider KE, Pickart RS, Lin P, Ashjian CJ and Chappell PD (2022) Coastal upwelling enhances abundance of a symbiotic diazotroph (UCYN-A) and its haptophyte host in the Arctic Ocean. Front. Mar. Sci. 9:877562. doi: 10.3389/fmars.2022.877562
Received: 16 February 2022; Accepted: 29 July 2022;
Published: 05 September 2022.
Edited by:
Jun Sun, China University of Geosciences Wuhan, ChinaReviewed by:
Matthew McGinness Mills, Stanford University, United StatesIrina N. Shilova, Second Genome, United States
Copyright © 2022 Selden, Einarsson, Lowry, Crider, Pickart, Lin, Ashjian and Chappell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Corday R. Selden, crselden@marine.rutgers.edu
†Present addresses: Corday R. Selden, Department of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ, United States
Sveinn V. Einarsson, Department of Microbiology and Cell Science, University of Florida, Gainesville, FL, United States
‡These authors have contributed equally to this work and share first authorship
 Corday R. Selden
Corday R. Selden Sveinn V. Einarsson
Sveinn V. Einarsson Kate E. Lowry
Kate E. Lowry Katherine E. Crider1
Katherine E. Crider1  Robert S. Pickart
Robert S. Pickart Peigen Lin
Peigen Lin Carin J. Ashjian
Carin J. Ashjian P. Dreux Chappell
P. Dreux Chappell